DEPDC1B
| DEPDC1B | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||
| Aliases | DEPDC1B, BRCC3, XTP1, DEP domain containing 1B | ||||||||||||||||
| External IDs | MGI: 2145425 HomoloGene: 10157 GeneCards: DEPDC1B | ||||||||||||||||
| |||||||||||||||||
| Orthologs | |||||||||||||||||
| Species | Human | Mouse | |||||||||||||||
| Entrez | |||||||||||||||||
| Ensembl | |||||||||||||||||
| UniProt | |||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||
| RefSeq (protein) | |||||||||||||||||
| Location (UCSC) | Chr 5: 60.6 – 60.7 Mb | Chr 13: 108.32 – 108.41 Mb | |||||||||||||||
| PubMed search | [1] | [2] | |||||||||||||||
| Wikidata | |||||||||||||||||
| View/Edit Human | View/Edit Mouse |
DEP Domain Containing Protein 1B also known as XTP1, XTP8,HBV XAg-Transactivated Protein 8, [formerly referred to as BRCC3] is a human protein encoded by a gene of similar name located on chromosome 5.[3][4][5]
The precise function of DEPDC1B is currently unknown. Expression profiles indicate that DEPDC1B is highly expressed ubiquitously throughout human tissue.[6]
Gene structure
Gene neighborhood
DEPDC1B is found on the long arm of chromosome 5 (5q12.1), spanning 103kb on the minus strand. The gene neighborhood of DEPDC1B includes 5 other genes. Downstream are two genes SEPT21 and PDE4D. Upstream are another two genes ELOV7 and KRT8P31. On the complement strand is another gene in the same region PART1.[4]
Promoter
DEPDC1B promoter region contains several transcription factors associated with proteins of ubiquitous expression. These transcription factors possess a central theme of cellular proliferation, cell cycle regulation, apoptosis, and differentiation. Few promoters unique to tumor suppression or tumorgenesis exist within the region as well.[7]
The following includes the top twenty Predicted Transcription Factors:
|
|
|
|
mRNA structure
Splice variants
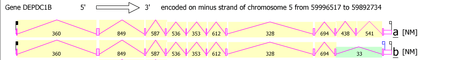
DEPDC1B possesses 13 mRNA splice variants that form two isoforms. Isoform 1 is the longest and is the most commonly used version of the gene. It is composed of 11 exons and is 103254bp in length. Isoform 2 is the second confirmed transcript variant. It is composed of 10 exons, missing the tenth exon of the first variant. The missing exon is 186bp in length.[8] See Protein Structure section for more detail...
Secondary structure
DEPDC1B is predicted to be predominantly alpha-helical. No significant beta-strands or beta structures exist with the protein. .[9]
Stem loops and binding miRNA
DEPDC1B is predicted to possess multiple stem loops in its 5' and 3' untranslated regions (UTR)[10][11] . In the 3' UTR, miRNA has-miR-499-5p binds to a nucleotide region predicted as a stem loop.[12]
Protein structure
Sequence
The DEPDC1B gene possesses two novel proteoforms. The longest variation, coded by mRNA isoform 1, is the most commonly used. The protein is 529 amino acids in length. The second novel proteoform, DEPDC1B.2 is coded by 10 exons, missing the 10th exon from the longest variation. The protein is 467 amino acids in length. The missing 62 amino acids follow the RhoGAP domain, in a region predicted to be highly phosphorylated[13]
Domains
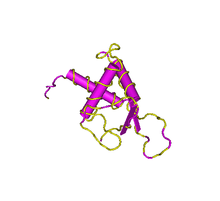
DEPDC1B contains two structural domains: a DEP domain and a RhoGAP domain.
The DEP domain is primarily found in proteins involved in G-protein signalling pathways and regulation of GTPase.[14][15] As well, experimental evidence suggests that the DEP domain determines the subcellular target of some GTPase Activating proteins.[16] In the DEPDC1B protein electronic inference has verified the GTPase activator activity function.[13] The solution structure of human containing DEP domain containing proteins verifies the secondary structure of the domain: containing three alpha-helices and two beta-strands within the approximate 80 amino acid region of the domain.[17][18]
The RhoGAP domain is another structural domain known to interact with GTPase and small GTPase. Research concerning the domain in other proteins indicates an approximately similar function among the domain in various proteins. The domain has been verified to interact with other proteins to form complexes or interact with other strcuutres of the cell such as the cytoskeleton or plasma membrane.[19]
Post-translational modification
DEPDC1B protein product is predicted to be highly phosphorylated after translation.[20] A single sumolaytion site, found within the RhoGAP domain, indicates the possible interaction of the protein with a SUMO protein, enabling or inhibiting interaction with other proteins.[21] A single palmitoylation site, found within the RhoGAP domain, indicates the possible interaction of the DEPDC1B protein product with a membrane via lipid anchor.[22]
No conserved glycolation sites are predicted within the mature DEPDC1B protein product.[23] No signal peptide or transmembrane domains are predicted within human or any ortholog protein.[24][25] No prenylation sites are predicted in any DEPDC1B orthologs.[26]
Expression
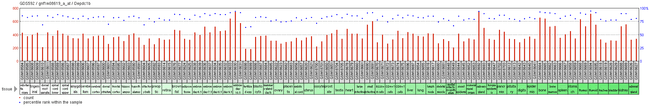
Expression of DEPDC1B is reported to be largely ubiquitous throughout mouse tissue. High level of gene expression is observed in all periods of life, except early zygote stages.[6] Experimental evidence suggests that DEPDC1B presents similar ubiquitous expression in all tissues.[27]
Differential expression profiles suggest that DEPDC1B is higher expressed in many cancerous disease states, including: papillary thyroid cancer,[28] breast cancer,[29] synovial sarcoma,[30] and prostatic cancer progression.[31] Also, DEPDC1B expression decreases in environments of beta-catenin depletion in multiple myeloma cell lines[32]
Interactions
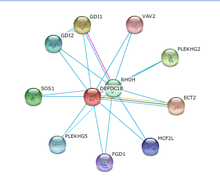
No interactions of DEPDC1B within any other protein product characterized by experimentation have been verified.[33]
Medium coexpression of DEPDC1B with ECT2 in cell cycle regulation and DNA synthesis was verified by similarity of expression in various tissues.[33] The remaining predicted interaction were determined via datamining.
Homology
Orthologs
DEPDC1B is unique to Chordates in Kingdom Animalia[34]
Multiple sequence alignments verify that DEPDC1B is highly conserved among orthologs.[35][36][37][38] The two structural domains (DEP and RhoGAP) are the two most conserved elements of the proteins. Various motifs are also conserved throughout the protein. No data suggesting motif function could be determined. All predicted post-translational modification were confirmed to be conserved in the orthologous proteins.
DEPDC1B evolution is predicted to follow the general species evolution.
| Genus and species | Common name | Class | Divergence (mya)[39] | Accession | Percent Identity[34] |
|---|---|---|---|---|---|
| Nomascus leucogenys | Northern white-cheeked gibbon | Mammalia | 20.4 | XP_003266016 | 98% |
| Papio anubis | Olive baboon | Mammalia | 29 | XP_003899752 | 98% |
| Mus musculus | House mouse | Mammalia | 92.3 | NP_848798 | 94% |
| Pteropus alecto | Black flying fox | Mammalia | 94.2 | XP_006906108 | 96% |
| Felis catus | Domestic cat | Mammalia | 94.2 | XP_003981045 | 96% |
| Bos taurus | Cow | Mammalia | 94.2 | XP_005221558 | 95% |
| Monodelphis domestica | Gray short-tailed opossum | Mammalia | 162.6 | XP_001363879 | 88% |
| Ficedula albicollis | Collared flycatcher | Ave | 296 | XP_005060715 | 77% |
| Taeniopygia guttata | Zebra finch | Ave | 296 | XP_002188294 | 76% |
| Gallus gallus | Chicken | Ave | 296 | NP_001006576 | 75% |
| Anolis carolinensis | Green anole | Reptilia | 296 | XP_003216290 | 76% |
| Xenopus tropicalis | Western clawed frog | Amphibia | 371.2 | NP_001121488 | 68% |
| Lepisosteus oculatus | Spotted gar | Actinopterygii | 400.1 | XP_006626875 | 68% |
| Maylandia zebra | Zebra mbuna | Actinopterygii | 400.1 | XP_004566850 | 57% |
Paralogs
.gif)
DEPDC1B possesses two significant paralogs - DEPDC1A and DEPDC7
Multiple sequence alignment and phylogenetic analysis indicates DEPDC1A as the most recent paralog, diverging approximately 600 million years ago. DEPDC1A has been researched in several disease states. High expression of the protein in Multiple Myeloma (MM) malignant plasma cells is associated with patient fatality. The high expression has been confirmed using conditional lentiviral vector delivery “to inhibit growth of human melanoma cell lines (HMCLs), with a block in G2 phase of the cell cycle, p53 phosphorylation and stabilization, and p21Cip1 accumulation”9.[40] In the same study it was concluded that DEPDC1A may contribute to the plasmablast features of MM cells, blocking differentiation. Study of DEPDC1A in bladder carcinogenesis revealed the gene as a possible antigen for the formation of bladder cancer cells. Using microarray and northern blotting confirmed the presence of unsubstantial amounts of the protein with in the normal tissues, excluding the testis. Currently the gene is a potential target molecule for therapeutic treatment of bladder carcinogenesis.[41]
No data detailing signigicant function in DEPD7 has been published or recorded.
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Morimoto, K (1996). "Characterization of a unique variant of bat rabies virus responsible for newly emerging human cases in North America". Proceedings of the National Academy of Sciences. 93 (11): 5653–5658. doi:10.1073/pnas.93.11.5653.
- 1 2 Entrez Gene: DEPDC1B http://www.ncbi.nlm.nih.gov/gene?LinkName=protein_gene&from_uid=223633999
- ↑ Gene Cards: DEPDC1B http://www.genecards.org/cgi-bin/carddisp.pl?gene=DEPDC1B
- 1 2 NCBI GEO http://www.ncbi.nlm.nih.gov/geoprofiles
- ↑ Genomatix Software. "Genomatix ElDorado". Retrieved 1998-2014.
- ↑ NCBI AceView http://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/av.cgi?db=human&q=DEPDC1B
- ↑ BPS : A. W. Burgess and P. K. Ponnuswamy and H. A. Sheraga, Analysis of conformations of amino acid residues and prediction of backbone topography in proteins, Israel J. Chem., p239-286, 1974, vol12. D_R : G. Dele`age and B. Roux, An algorithm for secondary structure prediction based on class prediction, Protein Engineering, p289-294, 1987, vol 1, num 4. DSC : Ross D. King and Michael J.E. Sternberg - Identification and application of the concepts important for accurate and reliable protein secondary structure prediction. Protein Science, 1996, 5:2298-2310 GGR : Garnier, Gibrat, and Robson, Meth. Enzym., R.F. Doolittle ed. 1996, 266:97-120 GOR : Jean Garnier and D. J. Osguthorpe and Barry Robson, Analysis of the accuracy and implications of simple methods for predicting the secondary structure of proteins, J. Mol. Biol., p 97-120, 1978, vol 120. G_G : O. Gascuel and J. L. Golmard, A simple method for predicting the secondary structure of globular proteins: implications and accuracy, CABIOS, p 357-365, 1988, vol 4. H_K : L. Howard Holley and Martin Karplus, Protein secondary structure prediction with a neural network, Proc. Natl. Acad. Sci. USA, p 152-156, Jan 1989, vol 86. K_S : Ross D. King and Michael J. E. Sternberg, Machine learning approach for the prediction of protein secondary structure, J. Mol. Biol., p 441-457, 1990, vol 216. L_G : Jonathan M. Levin and Jean Garnier, Improvements in a secondary structure prediction method based on a search for local sequence homologies and its use as a model building tool, Biochim. Biophys. Acta., p 283-295, 1988, vol 955. Q_S : Ning Qian and Terence Sejnowski, Predicting the secondary structure of proteins using neural network models, J. Mol. Biol., p 865-884, 1988, vol 202. JOI Joint prediction - Prediction made by the program that assigns the structure using a "winner takes all" procedure for each amino acid prediction using the other methods.
- ↑ Mfold http://mfold.rna.albany.edu/
- ↑ Sfold http://sfold.wadsworth.org/
- ↑ TragetScan http://www.targetscan.org/
- 1 2 Q8WUY9 (DEP1B_HUMAN) http://www.uniprot.org/uniprot/Q8WUY9
- ↑ Burchett, SA (2000). "Regulators of G protein signaling: a bestiary of modular protein binding domains". J. Neurochem. 75 (4): 1335–51. doi:10.1046/j.1471-4159.2000.0751335.x. PMID 10987813.
- ↑ Wong, HC; Mao, J; Nguyen, JT; Srinivas, S; Zhang, W; Liu, B; Li, L; Wu, D; Zheng, J (2000). "Structural basis of the recognition of the dishevelled DEP domain in the Wnt signaling pathway". Nat. Struct. Biol. 7 (12): 1178–84. doi:10.1038/82047. PMID 11101902.
- ↑ Martemyanov, K; et al. (2003). "The DEP Domain Determines Subcellular Targeting of the GTPase Activating Protein RGS9 In Vivo". The Journal of Neuroscience. 23 (12): 10175–10181.
- ↑ Zhang HP, Hayashi F, Yokoyama S. (2007) Solution structure of the dep domain from human dep domain-containing protein 1. http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=2ysr
- ↑ Madej T, Addess KJ, Fong JH, Geer LY, Geer RC, Lanczycki CJ, Liu C, Lu S, Marchler-Bauer A, Panchenko AR, Chen J, Thiessen PA, Wang Y, Zhang D, Bryant SH. "MMDB: 3D structures and macromolecular interactions." Nucleic Acids Res. 2012 Jan; 40(Database issue):D461-4
- ↑ Peck, J.; Douglas, G.; Wu, C. H.; Burbelo, P. D. (2002). "Human RhoGAP domain-containing proteins: structure, function and evolutionary relationships". FEBS Letters. 528 (1-3): 27–34. doi:10.1016/s0014-5793(02)03331-8. PMID 12297274.
- ↑ Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. Blom, N., Gammeltoft, S., and Brunak, S. Journal of Molecular Biology: 294(5): 1351-1362, 1999.
- ↑ Expasy SumoSP http://sumosp.biocuckoo.org/
- ↑ Expasy CSS-Palm http://csspalm.biocuckoo.org/
- ↑ Prediction of N-glycosylation sites in human proteins. R. Gupta, E. Jung and S. Brunak. In preparation, 2004.
- ↑ On filtering false positive transmembrane protein predictions": Miklos Cserzo, Frank Eisenhaber, Birgit Eisenhaber, and Istvan Simon; Protein Eng. 2002 15: 745-752
- ↑ SignalP 4.0: discriminating signal peptides from transmembrane regions Thomas Nordahl Petersen, Søren Brunak, Gunnar von Heijne & Henrik Nielsen Nature Methods, 8:785-786, 2011 doi: 10.1038/nmeth.1701 PMID 21959131
- ↑ Expasy PrePS http://mendel.imp.ac.at/sat/PrePS/index.html
- ↑ BioGPS http://biogps.org/#goto=genereport&id=55789
- ↑ NCBI GEO http://www.ncbi.nlm.nih.gov/geoprofiles/18885436
- ↑ NCBI GEO http://www.ncbi.nlm.nih.gov/geoprofiles/36185472M
- ↑ NCBI GEO http://www.ncbi.nlm.nih.gov/geoprofiles/38187695
- ↑ NCBI GEO http://www.ncbi.nlm.nih.gov/geoprofiles/14261636
- ↑ NCBI GEO http://www.ncbi.nlm.nih.gov/geoprofiles/61462636
- 1 2 String http://string-db.org/newstring_cgi/show_network_section.pl
- 1 2 NCBI BLAST http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ↑ Higgins, D.G.; Bleasby, A.J.; Fuchs, R. (1992). "CLUSTAL V: improved software for multiple sequence alignment". Computer Applications in the Biosciences (CABIOS). 8 (2): 189–191. doi:10.1093/bioinformatics/8.2.189. PMID 1591615.
- ↑ Thompson, J.D.; Higgins, D.G.; Gibson, T.J. (1994). "CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice". Nucleic Acids Res. 22: 4673–4680. doi:10.1093/nar/22.22.4673. PMC 308517
 . PMID 7984417.
. PMID 7984417. - ↑ Felsenstein, J (1989). "PHYLIP -- Phylogeny Inference Package (Version 3.2)". Cladistics. 5: 164–166.
- ↑ CLUSTAL W: Julie D. Thompson, Desmond G. Higgins and Toby J. Gibson, modified; any errors are due to the modifications. PHYLIP: Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle.
- ↑ Hedges, SB; Dudley, J; Kumar, S (2006). "TimeTree: a public knowledge-base of divergence times among organisms". Bioinformatics. 22: 2971–2972. doi:10.1093/bioinformatics/btl505. PMID 17021158.
- ↑ Kassambara, A., Schoenhals, M., & Klein, B. (2014). Inhibition of DEPDC1A , a Bad Prognostic Marker in Multiple Myeloma , Delays Growth and Induces Mature Plasma Cell Markers in Malignant Plasma Cells Results Increased Expression of DEPDC1A Gene in Multiple Myeloma Cells Compared to Normal Bone Marrow Plasma Cells in Association with a Poor Prognosis, 1–15.
- ↑ Kanehira, M.; Harada, Y.; Takata, R.; Shuin, T.; Miki, T.; Fujioka, T.; Katagiri, T. (2007). "Involvement of upregulation of DEPDC1 (DEP domain containing 1) in bladder carcinogenesis". Oncogene. 26 (44): 6448–55. doi:10.1038/sj.onc.1210466.