Morphobank
MorphoBank is a web application for collaborative evolutionary research, specifically phylogenetic systematics or cladistics, on the phenotype. Historically, scientists conducting research on phylogenetic systematics have worked individually or in small groups employing traditional single-user software applications such as MacClade,[1] Mesquite [2] and Nexus Data Editor.[3] As the hypotheses under study have grown more complex, large research teams have assembled to tackle the problem of discovering the tree of life for the estimated 4-100 million living species(Wilson 2003, pp. 77–80) and the many thousands more extinct species known from fossils. Because the phenotype is fundamentally visual, as phenotype-based phylogentic studies increase in size it becomes important that observations be backed up by labeled images. Traditional desktop software applications currently in wide use do not provide robust support for team-based research or for image manipulation and storage. MorphoBank is a particularly important tool for the growing scientific field of phenomics.
Advantages
Large phylogenetics research teams require simultaneous access by each member of the team to a single and secure copy of the team's data during a scientific research project. This single copy of the data also changes with great frequency during the data collection phase. Images that can be very helpful for documenting homology statements must be displayed, labeled and shared as homology statements develop. This cannot be accomplished elegantly with a desktop software package alone because in a desktop environment each collaborator is working on his own private copy of project data. Changes made by one participant cannot automatically propagate to others, preventing collaborators from seeing each other's data edits until they are manually (and due to the effort involved, often only periodically) merged into a single "true" dataset. In all but the smallest and most disciplined of teams, file version control and the reconciliation of changes made on multiple copies of the data emerge quickly as significant drags on productivity.
MorphoBank is an attempt to address these issues by leveraging the ubiquity of the web and modern web-based application techniques, including Ajax, web service layers, and rich internet applications to provide a full-featured, net-accessible collaborative workspace for phylogenetic research. In particular, MorphoBank makes it easy to:
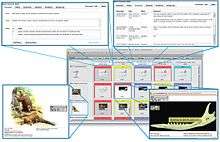
- Share all kinds of data with geographically separated team members, including taxonomy, character and specimen data, media (including images, video and audio), phylogenetic matrices (including data in the widely used NEXUS format) and other data such as documents and genetic sequences.
- Label high-resolution images using a web-based image annotation application.
- Collaboratively edit project data such as phylogenetic matrices using a built-in web-based matrix editor. The editor allows the linking of labeled images to individual cells of a matrix.
- Manage access to project data. Access ranges from full-access for team members to anonymous read-only access for potential reviewers.
- Publish completed project data on the web in support of a published paper with a persistent URL.
- Search The Encyclopedia of Life for taxon exemplar images.
- Create ontologies for updating and populating matrix cells.
These tasks are difficult or impossible in most existing software applications.
History
In 2001 the National Science Foundation sponsored a workshop,[4] at the American Museum of Natural History in New York to develop the outlines of a web-based system for collaborative, media-rich research tool. An application prototype presented at the workshop was later refined with feedback from the workshop and became MorphoBank version 1.0. A grant from the US National Oceanic and Atmospheric Administration funded further revisions resulting in version 2.0, released in 2005. Current support from the NSF is funding current feature enhancements to MorphoBank. MorphoBank is hosted by Stony Brook University and received back up support from the American Museum of Natural History. The current version is 3.0. Rationale for the software was described in the journal Cladistics.[5] MorphoBank has also received support from NESCENT and the San Diego Supercomputer Center.
The San Diego Supercomputer Center provides technical and hosting resources to the MorphoBank project.
Usage
MorphoBank hosts the products of peer-reviewed scientific research on phenotypes. An increasing volume of systematics data is "born digital" and MorphoBank is well suited to handle this type of material. In August 24, 2007, 62 active research projects were hosted by MorphoBank, as well as 6 completed (and published) projects. By March 2013 over 1,600 scientists and their students were registered users and the site had 208 publicly available projects with over 27,000 images. As of March 11, 2014, 919 active research projects are hosted by MorphoBank, 298 of which are completed (and published) projects. MorphoBank software has been used to assemble phylogenetic research on such groups as mammals,[6] from bats[7] to whales,[8][9] bivalve molluscs,[10] arachnids,[11] fossil plants[12] and living and extinct amniotes.[13] It has also been used more broadly in evolutionary and paleontological research to host curated images associated with published research on lacewing insects[14] geckos,[15][16] raptor birds,[17] dinosaurs,[18] frogs[19] and nematodes.[20]
MorphoBank has been particularly important to the Assembling the Tree of Life initiative sponsored by the National Science Foundation. MorphoBank is well-suited to such projects because of its tools for merging taxonomic, character and matrix-based data, as well as its collaborative features.[21] Highlights of this research include a collaborative matrix on mammal evolution published in Science that included over 4,000 phenomic characters scored for over 80 species,[22] a matrix on extant baleen whales featuring nearly 600 images,[23] and more.
References
- ↑ MacClade, a computer program for phylogenetic analysis, David R. Maddison and Wayne P. Maddison.
- ↑ Mesquite, A modular system for evolutionary analysis, Wayne P. Maddison
- ↑ Nexus Data Editor A program to edit NEXUS format data files, Roderick D. M. Page
- ↑ MorphoBank workshop report, November 10–11, 2001
- ↑ O'Leary, Maureen; Kaufman, Seth (October 2011). "MorphoBank: phylophenomics in the "cloud"". Cladistics. 27 (5): 529–537. doi:10.1111/j.1096-0031.2011.00355.x.
- ↑ O'Leary, M.A.; J.I. Bloch, J.J. Flynn, T.J. Gaudin, A. Giallombardo, N.P. Giannini, S.L. Goldber, B.P. Kraatz, Z.-X. Luo, J. Meng, X. Ni, M.J. Novacek, F.A. Perini, Z. Randall, G.W. Rougier, E.J. Sargis, M.T. Silcox, N.B. Simmons, M. Spaulding, P.M. Velazco, M. Weksler, J.R. Wible, and A.L. Cirranello (2013). "The placental mammal ancestor and the post-K-Pg radiation of placentals". Science. 332: 662–667. doi:10.1126/science.1229237. PMID 23393258. Cite uses deprecated parameter
|coauthors=(help) - ↑ "MorphoBank Project 265, Phylogenetic relationships of Icaronycteris, Archaeonycteris, Hassianycteris, and Palaeochiropteryx to extant bat lineages, with comments on the evolution of echolocation and foraging strategies in Microchiroptera".
- ↑ "MorphoBank Project 470, The Comparative Osteology of the Petrotympanic Complex (Ear Region) of Extant Baleen Whales (Cetacea: Mysticeti) Data Matrix".
- ↑ "MorphoBank Project 578, The pygmy right whale Caperea marginata - the last of the cetotheres".
- ↑ "MorphoBank Project 790, Investigating the Bivalve Tree of Life -- BivAToL matrix (100+ taxa)".
- ↑ "MorphoBank Project 44, New genus of cyphophthalmid from the Iberian Peninsula with a phylogenetic analysis of the Sironidae (Arachnida: Opiliones: Cyphophthalmi) and a SEM database of external morphology".
- ↑ "MorphoBank Project 277, Matrix of Morphological Characters of Humiriaceae".
- ↑ "MorphoBank Project 46, Integration of Morphological Data Sets for Phylogenetic Analysis of Amniota: The Importance of Integumentary Characters and Increased Taxonomic Sampling".
- ↑ "MorphoBank Project 146, A new genus and species of green lacewings from Brazil (Neuroptera: Chrysopidae: Leucochrysini)".
- ↑ "MorphoBank Project 348, Taxonomic revision of the cape verdean reptiles ii".
- ↑ "MorphoBank Project 1006, Two newly recognized species of Hemidactylus (Squamata, Gekkonidae) from the Arabian Peninsula and Sinai, Egypt".
- ↑ "MorphoBank Project 268, Predatory functional morphology in raptors: Interdigital variation in talon size is related to prey restraint and immobilisation technique".
- ↑ "MorphoBank Project 494, The ontogenetic osteohistology of Tenontosaurus tilletti".
- ↑ "MorphoBank Project 701, Phylogenetic signal and diversity of visceral pigmentation in eight anuran families".
- ↑ "MorphoBank Project 774, Cryptic species unveiled: the case of the nematode Spauligodon atlanticus.".
- ↑ The supermatrix approach to systematics, Alan de Queiroz and John Gatesy, TRENDS in Ecology and Evolution, Vol.22 No.1, 2006.
- ↑ O'Leary, M.A.; J.I. Bloch, J.J. Flynn, T.J. Gaudin, A. Giallombardo, N.P. Giannini, S.L. Goldber, B.P. Kraatz, Z.-X. Luo, J. Meng, X. Ni, M.J. Novacek, F.A. Perini, Z. Randall, G.W. Rougier, E.J. Sargis, M.T. Silcox, N.B. Simmons, M. Spaulding, P.M. Velazco, M. Weksler, J.R. Wible, and A.L. Cirranello (2013). "The placental mammal ancestor and the post-K-Pg radiation of placentals". Science. 332: 662–667. doi:10.1126/science.1229237. PMID 23393258. Cite uses deprecated parameter
|coauthors=(help) - ↑ Ekdale, E.G.; A. Berta; T.A. Deméré (2011). "The Comparative Osteology of the Petrotympanic Complex (Ear Region) of Extant Baleen Whales (Cetacea: Mysticeti)". PLoS ONE. 6 (6): e21311. doi:10.1371/journal.pone.0021311.
Citations
Wilson, E. O. (2003), "The encyclopedia of life", TREE, 18: 77–80.
External links
- MorphoBank home page
- Modernizing the Tree of Life, Science, 10 June 2003, 300: 1692-1697. Article discussing efforts of projects including MorphoBank to simplify and speed up the assessment of biodiversity.
- "Morphology: The Shape of Things to Come", Paul D. Thacker, BioScience, June 2003, Vol. 53 No. 6, 544. A report on contemporary initiatives in morphological research, including MorphoBank.