Phi X 174
| Phi X 174 | |
|---|---|
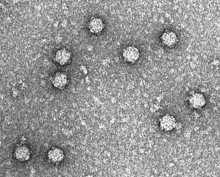 | |
| Electron micrograph of phage ΦX174 | |
| Virus classification | |
| Group: | Group II (ssDNA) |
| Family: | Microviridae |
| Genus: | Microvirus |
| Species: | Enterobacteria phage phiX174 |
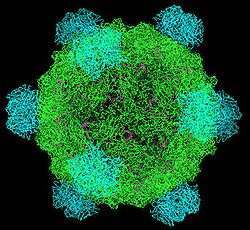
The phi X 174 (or ΦX174) bacteriophage is a virus and was the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977.[2] In 1962, Walter Fiers and Robert Sinsheimer had already demonstrated the physical, covalently closed circularity of ΦX174 DNA.[3] Nobel prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology.[4][5] In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides.[6] The ΦX174 virus particle has also been successfully assembled in vitro.[7] Recently, it was shown how its highly overlapping genome can be fully decompressed and still remain functional.[8]
Virology
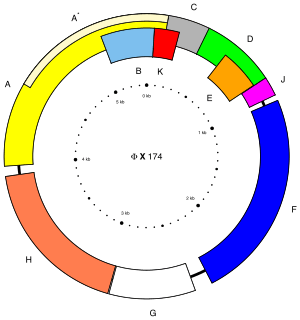
This bacteriophage has a [+] circular single-stranded DNA genome of 5386 nucleotides encoding 11 proteins.[1] Of these 11 genes, only 8 are essential to viral morphogenesis. The GC-content is 44% and 95% of nucleotides belong to coding genes.
| Protein | Copies | Function[9] |
|---|---|---|
| A | -- | Nicks RF DNA to initiate rolling-circle replication; ligates ends of linear phage DNA to form single-stranded circular DNA |
| A* | -- | Inhibits host cell DNA replication; blocks superinfecting phage; not essential |
| B | 60 in procapsid | Internal scaffolding protein involved in procapsid assembly |
| C | -- | DNA packaging |
| D | 240 in procapsid | External scaffolding protein involved in procapsid assembly |
| E | -- | Host cell lysis |
| F | 60 in virion | Major capsid protein |
| G | 60 in virion | Major spike protein |
| H | 12 in virion | DNA pilot protein (or minor spike protein) |
| J | 60 in virion | Binds to new single-stranded phage DNA; accompanies phage DNA into procapsid |
| K | -- | Optimizes burst size; not essential |
Infection begins when G protein binds to lipopolysaccharides on the bacterial host cell surface. H protein (or the DNA Pilot Protein) pilots the viral genome through the bacterial membrane of E.coli bacteria (Jazwinski et al. 1975) most likely via a predicted N-terminal transmembrane domain helix (Tusnady and Simon, 2001). However, it has become apparent that H protein is a multifunctional protein (Cherwa, Young and Fane, 2011). This is the only viral capsid protein of ΦX174 to lack a crystal structure for a couple of reasons. It has low aromatic content and high glycine content, making the protein structure very flexible and in addition, individual hydrogen atoms (the R group for glycines) are difficult to detect in protein crystallography. Additionally, H protein induces lysis of the bacterial host at high concentrations as the predicted N-terminal transmembrane helix easily pokes holes through the bacterial wall. By bioinformatics, this protein contains four predicted coiled-coil domains which has a significant homology to known transcription factors. Additionally, it was determined by Ruboyianes et al. (2009) that de novo H protein was required for optimal synthesis of other viral proteins. Interestingly, mutations in H protein that prevent viral incorporation, can be overcome when excess amounts of Protein B, the internal scaffolding protein, are supplied.
The DNA is ejected through a hydrophilic channel at the 5-fold vertex (McKenna et al. 1992). It is understood that H protein resides in this area but experimental evidence has not verified its exact location. Once inside the host bacterium, replication of the [+] ssDNA genome proceeds via negative sense DNA intermediate. This is done as the phage genome supercoils and the secondary structure formed by such supercoiling attracts a primosome protein complex. This translocates once around the genome and synthesizes a [-]ssDNA from the positive original genome. [+]ssDNA genomes to package into viruses are created from this by a rolling circle mechanism. This is the mechanism by which the double stranded supercoiled genome is nicked on the negative strand by a virus-encoded A protein, also attracting a bacterial DNA Polymerase to the site of cleavage. DNAP will use the negative strand as a template to make positive sense DNA. As it translocates around the genome it displaces the outer strand of already-synthesised DNA, which is immediately coated by ssBP proteins. The A protein will cleave the complete genome every time it recognises the origin sequence.
As D protein is the most abundant gene transcript, it is the most protein in the viral procapsid. Similarly, gene transcripts for F, J, and G are more abundant than for H as the stoichiometry for these structural proteins is 5:5:5:1.The primosome are protein complexes which attach/bind the enzyme helicase on the template. primosomes gives RNA primers for DNA synthesis to strands.
Notes
Phi X is regularly used as a positive control in DNA sequencing due to its relatively small genome size in comparison to other organisms and the extensive work that has been done on it.
See also
References
- 1 2 Enterobacteria phage phiX174 sensu lato, complete genome. "Complete genome: accession NC_001422", National Center for Biotechnology Information. Retrieved on 30 January 2016.
- ↑ Sanger, F.; Air, G. M.; Barrell, B. G.; Brown, N. L.; Coulson, A. R.; Fiddes, J. C.; Hutchison, C. A.; Slocombe, P. M.; Smith, M. (1977). "Nucleotide sequence of bacteriophage ΦX174 DNA". Nature. 265 (5596): 687–95. Bibcode:1977Natur.265..687S. doi:10.1038/265687a0. PMID 870828.
- ↑ Fiers, Walter; Sinsheimer, Robert L. (1962). "The structure of the DNA of bacteriophage ΦX174". Journal of Molecular Biology. 5 (4): 424. doi:10.1016/S0022-2836(62)80031-X.
- ↑ National Library of Medicine Profiles in Science. The Arthur Kornberg Papers. "Creating Life in the Test Tube," 1959-1970. link
- ↑ Goulian, Mehran; Kornberg, Arthur; Sinsheimer, Robert L. (1967). "Enzymatic Synthesis of DNA, XXIV. Synthesis of Infectious Phage ΦX174 DNA". Proceedings of the National Academy of Sciences. 58 (6): 2321–2328. Bibcode:1967PNAS...58.2321G. doi:10.1073/pnas.58.6.2321. JSTOR 58720. PMC 223838
 . PMID 4873588.
. PMID 4873588. - ↑ Smith, Hamilton O.; Hutchison, Clyde A.; Pfannkoch, Cynthia; Venter, J. Craig (2003). "Generating a Synthetic Genome by Whole Genome Assembly: ΦX174 Bacteriophage from Synthetic Oligonucleotides". Proceedings of the National Academy of Sciences. 100 (26): 15440–5. Bibcode:2003PNAS..10015440S. doi:10.1073/pnas.2237126100. JSTOR 3149024. PMC 307586
 . PMID 14657399.
. PMID 14657399. - ↑ Cherwa, James E.; Organtini, Lindsey J.; Ashley, Robert E.; Hafenstein, Susan L.; Fane, Bentley A. (2011). "In Vitro Assembly of the ΦX174 Procapsid from External Scaffolding Protein Oligomers and Early Pentameric Assembly Intermediates". Journal of Molecular Biology. 412 (3): 387–96. doi:10.1016/j.jmb.2011.07.070. PMID 21840317.
- ↑ Jaschke, Paul R.; Lieberman, Erica K.; Rodriguez, Jon; Sierra, Adrian; Endy, Drew (2012). "A fully decompressed synthetic bacteriophage ΦX174 genome assembled and archived in yeast". Virology. 434 (2): 278–84. doi:10.1016/j.virol.2012.09.020. PMID 23079106.
- ↑ Fane, Bentley A.; Brentlinger, Karie L.; Burch, April D.; Chen, Min; Hafenstein, Susan; Moore, Erica; Novak, Christopher R.; Uchiyama, Asako (2006). "ɸX174 et al., the Microviridae". In Calender, Richard. The Bacteriophages (2nd ed.). New York: Oxford Univ. Press. p. 130. ISBN 978-0195148503.
External links
- Goodsell, David (February 2000). "Bacteriophage phiX174". Molecule of the Month. RCSB-PDB.