Shotgun sequencing
In genetics, shotgun sequencing, also known as shotgun cloning, is a method used for sequencing long DNA strands. It is named by analogy with the rapidly expanding, quasi-random firing pattern of a shotgun.
The chain termination method of DNA sequencing (or "Sanger sequencing" for its developer Frederick Sanger) can only be used for fairly short strands of 100 to 1000 base pairs. Longer sequences are subdivided into smaller fragments that can be sequenced separately, and subsequently they are re-assembled to give the overall sequence. Two principal methods are used for this: primer walking (or "chromosome walking") which progresses through the entire strand piece by piece, and shotgun sequencing, which is a faster but more complex process that uses random fragments.
In shotgun sequencing,[1][2] DNA is broken up randomly into numerous small segments, which are sequenced using the chain termination method to obtain reads. Multiple overlapping reads for the target DNA are obtained by performing several rounds of this fragmentation and sequencing. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence.[1]
Shotgun sequencing was one of the precursor technologies that was responsible for enabling full genome sequencing.
Example
For example, consider the following two rounds of shotgun reads:
| Strand | Sequence |
|---|---|
| Original | AGCATGCTGCAGTCATGCTTAGGCTA |
| First shotgun sequence | AGCATGCTGCAGTCATGCT--------------------------TAGGCTA |
| Second shotgun sequence | AGCATG--------------------------CTGCAGTCATGCTTAGGCTA |
| Reconstruction | AGCATGCTGCAGTCATGCTTAGGCTA |
In this extremely simplified example, none of the reads cover the full length of the original sequence, but the four reads can be assembled into the original sequence using the overlap of their ends to align and order them. In reality, this process uses enormous amounts of information that are rife with ambiguities and sequencing errors. Assembly of complex genomes is additionally complicated by the great abundance of repetitive sequences, meaning similar short reads could come from completely different parts of the sequence.
Many overlapping reads for each segment of the original DNA are necessary to overcome these difficulties and accurately assemble the sequence. For example, to complete the Human Genome Project, most of the human genome was sequenced at 12X or greater coverage; that is, each base in the final sequence was present on average in 12 different reads. Even so, current methods have failed to isolate or assemble reliable sequence for approximately 1% of the (euchromatic) human genome, as of 2004.[3]
Whole genome shotgun sequencing
History
The first genome sequenced by shotgun sequencing was that of cauliflower mosaic virus, published in 1981.[4][5] However, whole genome shotgun sequencing for small (4000- to 7000-base-pair) genomes had been suggested already in 1979.[1]
Paired-end sequencing
Broader application benefited from pairwise end sequencing, known colloquially as double-barrel shotgun sequencing. As sequencing projects began to take on longer and more complicated DNA sequences, multiple groups began to realize that useful information could be obtained by sequencing both ends of a fragment of DNA. Although sequencing both ends of the same fragment and keeping track of the paired data was more cumbersome than sequencing a single end of two distinct fragments, the knowledge that the two sequences were oriented in opposite directions and were about the length of a fragment apart from each other was valuable in reconstructing the sequence of the original target fragment.
History. The first published description of the use of paired ends was in 1990[6] as part of the sequencing of the human HGPRT locus, although the use of paired ends was limited to closing gaps after the application of a traditional shotgun sequencing approach. The first theoretical description of a pure pairwise end sequencing strategy, assuming fragments of constant length, was in 1991.[7] At the time, there was community consensus that the optimal fragment length for pairwise end sequencing would be three times the sequence read length. In 1995 Roach et al.[8] introduced the innovation of using fragments of varying sizes, and demonstrated that a pure pairwise end-sequencing strategy would be possible on large targets. The strategy was subsequently adopted by The Institute for Genomic Research (TIGR) to sequence the genome of the bacterium Haemophilus influenzae in 1995,[9] and then by Celera Genomics to sequence the Drosophila melanogaster (fruit fly) genome in 2000,[10] and subsequently the human genome.
Approach
To apply the strategy, a high-molecular-weight DNA strand is sheared into random fragments, size-selected (usually 2, 10, 50, and 150 kb), and cloned into an appropriate vector. The clones are then sequenced from both ends using the chain termination method yielding two short sequences. Each sequence is called an end-read or read and two reads from the same clone are referred to as mate pairs. Since the chain termination method usually can only produce reads between 500 and 1000 bases long, in all but the smallest clones, mate pairs will rarely overlap.
Assembly
The original sequence is reconstructed from the reads using sequence assembly software. First, overlapping reads are collected into longer composite sequences known as contigs. Contigs can be linked together into scaffolds by following connections between mate pairs. The distance between contigs can be inferred from the mate pair positions if the average fragment length of the library is known and has a narrow window of deviation. Depending on the size of the gap between contigs, different techniques can be used to find the sequence in the gaps. If the gap is small (5-20kb) then the use of PCR to amplify the region is required, followed by sequencing. If the gap is large (>20kb) then the large fragment is cloned in special vectors such as BAC (Bacterial artificial chromosomes) followed by sequencing of the vector.
Pros and cons
Proponents of this approach argue that it is possible to sequence the whole genome at once using large arrays of sequencers, which makes the whole process much more efficient than more traditional approaches. Detractors argue that although the technique quickly sequences large regions of DNA, its ability to correctly link these regions is suspect, particularly for genomes with repeating regions. As sequence assembly programs become more sophisticated and computing power becomes cheaper, it may be possible to overcome this limitation.
Coverage
Coverage (read depth or depth) is the average number of reads representing a given nucleotide in the reconstructed sequence. It can be calculated from the length of the original genome (G), the number of reads(N), and the average read length(L) as . For example, a hypothetical genome with 2,000 base pairs reconstructed from 8 reads with an average length of 500 nucleotides will have 2x redundancy. This parameter also enables one to estimate other quantities, such as the percentage of the genome covered by reads (sometimes also called coverage). A high coverage in shotgun sequencing is desired because it can overcome errors in base calling and assembly. The subject of DNA sequencing theory addresses the relationships of such quantities.
Sometimes a distinction is made between sequence coverage and physical coverage. Sequence coverage is the average number of times a base is read (as described above). Physical coverage is the average number of times a base is read or spanned by mate paired reads.[11]
Hierarchical Shotgun sequencing
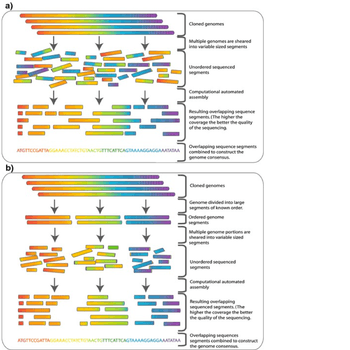
Although shotgun sequencing can in theory be applied to a genome of any size, its direct application to the sequencing of large genomes (for instance, the Human Genome) was limited until the late 1990s, when technological advances made practical the handling of the vast quantities of complex data involved in the process.[12] Historically, full-genome shotgun sequencing was believed to be limited by both the sheer size of large genomes and by the complexity added by the high percentage of repetitive DNA (greater than 50% for the human genome) present in large genomes.[13] It was not widely accepted that a full-genome shotgun sequence of a large genome would provide reliable data. For these reasons, other strategies that lowered the computational load of sequence assembly had to be utilized before shotgun sequencing was performed.[13] In hierarchical sequencing, also known as top-down sequencing, a low-resolution physical map of the genome is made prior to actual sequencing. From this map, a minimal number of fragments that cover the entire chromosome are selected for sequencing.[14] In this way, the minimum amount of high-throughput sequencing and assembly is required.
The amplified genome is first sheared into larger pieces (50-200kb) and cloned into a bacterial host using BACs or PACs. Because multiple genome copies have been sheared at random, the fragments contained in these clones have different ends, and with enough coverage (see section above) finding a scaffold of BAC contigs that covers the entire genome is theoretically possible. This scaffold is called a tiling path.
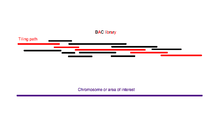
Once a tiling path has been found, the BACs that form this path are sheared at random into smaller fragments and can be sequenced using the shotgun method on a smaller scale.
Although the full sequences of the BAC contigs is not known, their orientations relative to one another are known. There are several methods for deducing this order and selecting the BACs that make up a tiling path. The general strategy involves identifying the positions of the clones relative to one another and then selecting the least number of clones required to form a contiguous scaffold that covers the entire area of interest. The order of the clones is deduced by determining the way in which they overlap.[15] Overlapping clones can be identified in several ways. A small radioactively or chemically labeled probe containing a sequence-tagged site (STS) can be hybridized onto a microarray upon which the clones are printed.[15] In this way, all the clones that contain a particular sequence in the genome are identified. The end of one of these clones can then be sequenced to yield a new probe and the process repeated in a method called chromosome walking.
Alternatively, the BAC library can be restriction-digested. Two clones that have several fragment sizes in common are inferred to overlap because they contain multiple similarly spaced restriction sites in common.[15] This method of genomic mapping is called restriction fingerprinting because it identifies a set of restriction sites contained in each clone. Once the overlap between the clones has been found and their order relative to the genome known, a scaffold of a minimal subset of these contigs that covers the entire genome is shotgun-sequenced.[14]
Because it involves first creating a low-resolution map of the genome, hierarchical shotgun sequencing is slower than whole-genome shotgun sequencing, but relies less heavily on computer algorithms than whole-genome shotgun sequencing. The process of extensive BAC library creation and tiling path selection, however, make hierarchical shotgun sequencing slow and labor-intensive. Now that the technology is available and the reliability of the data demonstrated,[13] and the speed and cost efficiency of whole-genome shotgun sequencing has made it the primary method for genome sequencing.
Shotgun and Next-generation sequencing
The classical shotgun sequencing was based on the Sanger sequencing method: this was the most advanced technique for sequencing genomes from about 1995–2005. The shotgun strategy is still applied today, however using other sequencing technologies, called next-generation sequencing. These technologies produce shorter reads (anywhere from 25–500bp) but many hundreds of thousands or millions of reads in a relatively short time (on the order of a day).[16] This results in high coverage, but the assembly process is much more computationally intensive. These technologies are vastly superior to Sanger sequencing due to the high volume of data and the relatively short time it takes to sequence a whole genome.[17]
See also
References
- 1 2 3 Staden, R (1979). "A strategy of DNA sequencing employing computer programs". Nucleic Acids Research. 6 (7): 2601–10. doi:10.1093/nar/6.7.2601. PMC 327874
 . PMID 461197.
. PMID 461197. - ↑ Anderson, S (1981). "Shotgun DNA sequencing using cloned DNase I-generated fragments". Nucleic Acids Research. 9 (13): 3015–27. doi:10.1093/nar/9.13.3015. PMC 327328
 . PMID 6269069.
. PMID 6269069. - ↑ Human Genome Sequencing Consortium, International (21 October 2004). "Finishing the euchromatic sequence of the human genome". Nature. 431 (7011): 931–945. doi:10.1038/nature03001. PMID 15496913.
- ↑ Gardner, Richard C.; Howarth, Alan J.; Hahn, Peter; Brown-Luedi, Marianne; Shepherd, Robert J.; Messing, Joachim (1981-06-25). "The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by M13mp7 shotgun sequencing". Nucleic Acids Research. 9 (12): 2871–2888. doi:10.1093/nar/9.12.2871. ISSN 0305-1048. PMC 326899
 . PMID 6269062.
. PMID 6269062. - ↑ Doctrow, Brian (2016-07-19). "Profile of Joachim Messing". Proceedings of the National Academy of Sciences. 113 (29): 7935–7937. doi:10.1073/pnas.1608857113. ISSN 0027-8424. PMID 27382176.
- ↑ Edwards, A; Caskey, T (1991). "Closure strategies for random DNA sequencing". Methods: A Companion to Methods in Enzymology. 3 (1): 41–47. doi:10.1016/S1046-2023(05)80162-8.
- ↑ Edwards, A; Voss, H.; Rice, P.; Civitello, A.; Stegemann, J.; Schwager, C.; Zimmerman, J.; Erfle, H.; Caskey, T.; Ansorge, W. (1990). "Automated DNA sequencing of the human HPRT locus". Genomics. 6 (4): 593–608. doi:10.1016/0888-7543(90)90493-E. PMID 2341149.
- ↑ Roach, JC; Boysen, C; Wang, K; Hood, L (1995). "Pairwise end sequencing: a unified approach to genomic mapping and sequencing". Genomics. 26 (2): 345–353. doi:10.1016/0888-7543(95)80219-C. PMID 7601461.
- ↑ Fleischmann, RD; et al. (1995). "Whole-genome random sequencing and assembly of Haemophilus influenzae Rd". Science. 269 (5223): 496–512. Bibcode:1995Sci...269..496F. doi:10.1126/science.7542800. PMID 7542800.
- ↑ Adams, MD; et al. (2000). "The genome sequence of Drosophila melanogaster". Science. 287 (5461): 2185–95. Bibcode:2000Sci...287.2185.. doi:10.1126/science.287.5461.2185. PMID 10731132.
- ↑ Meyerson, M.; Gabriel, S.; Getz, G. (2010). "Advances in understanding cancer genomes through second-generation sequencing". Nature Reviews Genetics. 11 (10): 685–696. doi:10.1038/nrg2841. PMID 20847746.
- ↑ Dunham, I. Genome Sequencing. Encyclopedia of Life Sciences, 2005. doi:10.1038/npg.els.0005378
- 1 2 3 Venter, J. C. ‘’Shotgunning the Human Genome: A Personal View.’’ Encyclopedia of Life Sciences, 2006.
- 1 2 Gibson, G. and Muse, S. V. A Primer of Genome Science. 3rd ed. P.84
- 1 2 3 Dear, P. H. Genome Mapping. Encyclopedia of Life Sciences, 2005. doi:10.1038/npg.els.0005353.
- ↑ Karl, V; et al. (2009). "Next Generation Sequencing: From Basic Research to Diagnostics". Clinical Chemistry. 55 (4): 41–47. doi:10.1373/clinchem.2008.112789. PMID 19246620.
- ↑ Metzker, Michael L. (2010). "Sequencing technologies - the next generation". Nat Rev Genet. 11 (1): 31–46. doi:10.1038/nrg2626. PMID 19997069.
Further reading
- "Shotgun sequencing comes of age". The Scientist. Retrieved December 31, 2002.
- "Shotgun sequencing finds nanoorganisms - Probe of acid mine drainage turns up unsuspected virus-sized Archaea". SpaceRef.com. Retrieved December 23, 2006.
- "Genomic shotgun sequencing". biology science. Retrieved April 11, 2009.
External links
![]() This article incorporates public domain material from the National Center for Biotechnology Information document "NCBI Handbook".
This article incorporates public domain material from the National Center for Biotechnology Information document "NCBI Handbook".