Spherical nucleic acid
Spherical nucleic acids (SNAs)[1] – defined as structures which are an arrangement of densely packed, highly oriented nucleic acids in a spherical geometry – were first introduced in 1996[2] by the Mirkin group at Northwestern University. The unique arrangement and orientation of one-dimensional linear nucleic acids within this three-dimensional framework results in fundamentally new chemical, biological, and physical properties, which represent a paradigm shift in the use of nucleic acids for intracellular gene regulation, molecular diagnostics, and materials synthesis applications.
Structure and Function
The first SNA consisted of 3’ alkanethiol-terminated, single-stranded oligonucleotides (short DNA sequences) covalently attached to the surface of spherical gold nanoparticles. A dense nucleic acid loading was achieved through a series of salt additions, in which positively charged counterions were used to reduce electrostatic repulsion between adjacent negatively charged DNA strands and thereby enable more efficient DNA packing onto the nanoparticle surface.
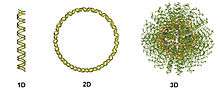
One- and two-dimensional forms of nucleic acids (e.g., single strands, linear duplexes, and plasmids) are important biological machinery for the storage and transmission of genetic information (Fig. 1). Underlying this utility are the bioprogrammable interactions between complementary nucleotide bases. Scientists and engineers have been synthesizing and, in certain cases, mass-producing such structures for decades in an effort to understand and exploit this elegant recognition motif. The recognition abilities of nucleic acids are enhanced when arranged in a spherical geometry, which allows for polyvalent interactions. This polyvalency, along with the high density and degree of orientation described above, helps to explain why a three-dimensional nucleic acid structure fundamentally composed of linear, one-dimensional nucleic acids exhibits dramatically different properties than its lower-dimensional constituents (Fig. 2).

Two decades of research on SNAs have revealed that they exhibit a synergistic combination of properties resulting from the inorganic core and the nucleic acid shell. The inorganic nanoparticle core (in addition to gold, silver,[3] iron oxide,[4] silica,[5] and semiconductor materials[6] have also been used) serves two purposes: 1) it imparts upon the conjugate novel physical and chemical properties (e.g. plasmonic, catalytic,[7][8] magnetic,[4] luminescent[6]), and 2) it acts as a scaffold for the assembly and orientation of nucleic acids into the dense arrangement that gives rise to many of their functional properties. The nucleic acid shell gives the conjugate unique, programmable chemical and biological recognition abilities that offer greater binding strengths and higher duplex stabilities compared to the same sequence of linear DNA,[7][9][10] cooperative melting behavior with DNA strands of a complementary sequence, and enhanced cellular uptake without the use of transfection agents.[11] Recent studies have shown that one can crosslink the DNA strands at their base, and subsequently dissolve the inorganic core with KCN or I2 to create a new coreless form of SNA (Fig. 3, right),[12] which exhibits many of the hallmark properties of the original DNA-nanoparticle conjugate (Fig. 3, left). This work underscores one of the fundamental features of SNAs: the properties of these nanomaterials are core-independent, derived from the orientation and packing of the nucleic acid shell.

Due to their unique structure and function, SNAs occupy a materials space distinct from “DNA nanotechnology and origami.”[13][14][15] With DNA origami, such structures are synthesized via DNA hybridization events. In contrast, the SNA structure can be synthesized independent of nucleic acid sequence and hybridization, instead relying upon robust chemical bond formation between nanoparticles and nucleic acid adsorbates. Furthermore, DNA origami uses DNA hybridization interactions to realize a final structure, whereas SNAs and other forms of three-dimensional nucleic acids (structures templated with a triangular prism, rod, octahedra, or rhombic dodecadhedra-shaped nanoparticles)[16] utilize the nanoparticle core to arrange the linear nucleic acid components into functional forms. Indeed, it is the particle core that dictates the shape of the SNA, and to date, single-stranded and double-stranded versions of these materials have been created that consist of DNA, LNA, and RNA.
SNAs should also not be confused with their monovalent analogues – individual particles coupled to a single DNA strand.[17] Such single strand-nanoparticle conjugate structures have led to interesting advances in their own right, but do not exhibit the unique properties of SNAs.
Applications and Societal Benefit
Intracellular Gene Regulation

SNAs also have advanced the field of nanotherapeutics as extraordinarily useful gene regulation agents.[12] Despite their high negative charge, these nanostructures are naturally taken up by cells, without the need for positively charged polymeric co-carriers, and are highly effective as gene regulation agents in both siRNA and antisense gene regulation pathways.[12][18] This discovery represents a paradigm shift for the field, since previous lore predicted that nucleic acids would not efficiently enter cells without positively charged co-carriers (Fig. 4). Although this is true for linear nucleic acids, it is not the case for SNAs. This stunning result is because SNAs, unlike their linear counterparts, have the ability to complex scavenger proteins and thereby facilitate endocytosis. Investigation of the mechanism for SNA uptake[19] revealed that these nanostructures exhibit an ability to deactivate enzymes responsible for triggering cellular immune response and oligonucleotide degradation.[20][21][22] These materials constitute a powerful single-entity gene regulation platform for intracellular biochemical and biological applications and represent therapeutic lead structures for a wide variety of oncology treatments. Indeed, the first therapeutic versions of these structures are currently entering clinical trials.
SNAs packed gold nanoparticles are able to deliver small interfering RNA (siRNA) to treat glioblastoma multiforme in a proof-of-concept study using mouse model, reported by the research team led by Mirkin. The iRNA targets Bcl2Like12 gene overexpressed in glioblastoma tumors and silences the oncogene. The gold nanoparticles injected intravenously cross the blood-brain barrier and naturally find their target in the brain. In the animal model, the treatment caused a 20% increase in survival rate and 3 to 4 fold reduction in tumor size. Mirkin indicated that this novel therapeutic approach establishes a platform for treating a wide range of diseases.[23][24]
Molecular Diagnostics

Spherical nucleic acids have catalyzed world-wide interest in using well-characterized nanostructures as novel labels for in vitro biodetection schemes and intracellular assays, and as potent cell transfection, therapeutic, and gene regulation materials.[12][25][26][27][28][29][30][31] SNAs have enabled the first commercialized molecular medical diagnostic systems of the modern era of nanotechnology (Fig. 5). The FDA-cleared Verigene System, commercialized by Nanosphere, is now sold in over twenty countries. This technology allows the detection of markers for many diseases, including infectious disease and cancers, with a sensitivity and selectivity far exceeding that of conventional diagnostic tools. Indeed, the Verigene is transforming patient care by transitioning molecular diagnostic screening from centralized, often remote, analytical laboratories to the local hospital setting, which dramatically decreases the time required for diagnosis. Further, the Verigene has enabled the identification of new markers for Alzheimer's disease,[26] HIV,[32][33] and cardiac disease[34][35][36] as well as new tests for the early detection of a variety of forms of prostate cancer.[37] These medical diagnostic and therapeutic tools have already saved or improved many lives. As such, SNAs represent an important class of diagnostic probes that will enable fundamental discoveries and empower physicians to make quick and accurate decisions about patient care regarding almost any disease with a genetic basis.
Intracellular Probes
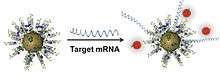
NanoFlares utilize the SNA architecture for intracellular mRNA detection. In this design, alkanethiol-terminated antisense DNA strands (complementary to a target mRNA strand within cells) are attached to the surface of a gold nanoparticle. Fluorophore-labeled “reporter strands” are then hybridized to the SNA construct to form the NanoFlare. When the fluorophore labels are brought within close proximity of the gold surface, as controlled by programmable nucleic acid hybridization, their fluorescence is quenched (Fig. 6).[30] Following cellular entry (enabled by the SNA geometry), reporter strands are concomitantly driven from the NanoFlare and replaced by the longer, target mRNA sequence. Note that mRNA binding is thermodynamically favored since the strands holding the reporter sequence have greater overlap of their nucleotide sequence with the target mRNA. Upon reporter strand release, dye fluorescence is no longer quenched by the gold nanoparticle core, and increased fluorescence is observed. This method for RNA detection provides the only way to sort live cells based upon genetic content, and has thus enabled significant advances in disease diagnosis and cellular genetics. AuraSense and AuraSense Therapeutics are two start-up companies founded to advance these SNA structures in several sectors of the biomedical industry. In 2011, AuraSense entered into partnership with EMD-Millipore to commercialize NanoFlares under the SmartFlare trade name. There are now over 1600 commercial forms of SmartFlares sold world-wide.
Materials Synthesis
SNAs also have been utilized to develop an entirely new field of materials science – one that focuses on using SNAs as synthetically-programmable building blocks in the construction of DNA-nanoparticle superlattices (Fig. 7). In 2011, a landmark paper was published in Science that defines a set of design rules for making superlattice structures of tailorable crystallographic symmetry and lattice parameters with sub-nm precision.[38] The rules are described as follows:

1. When all DNA-NPs in a system possess equal hydrodynamic radii, each NP in the thermodynamic product will maximize the number of nearest neighbors to which it can form DNA connections.
2. When two lattices are of similar stability, the kinetic product can be produced by slowing the rate at which individual DNA linkers dehybridize and subsequently rehybridize.
3. The overall hydrodynamic radius of a DNA-NP, rather than the sizes of its individual NP or oligonucleotide components, dicates its assembly and packing behavior.
4. In a binary system, the size ratio and DNA linker ratio between two particles dictate the thermodynamically favored crystal structure.
5. Two systems with the same size ratio and DNA linker ratio exhibit the same thermodynamic product.
6. The most stable crystal structure will maximize all possible types of DNA sequence-specific hybridization interactions.
These design rules are analogous to Pauling's rules for ionic crystals, but ultimately more powerful. For example, when using atomic or ionic building blocks in the construction of materials, the crystal structure, symmetry, and spacing are fixed by atomic radii and electronegativity. However, in the nanoparticle-based system, crystal structure can be tuned independent of the nanoparticle size and composition by simply adjusting the length and sequence of the attached DNA. This has enabled the construction of novel crystal structures for several materials systems and even crystal structures with no mineral equivalents.[39] The ability to place nanoparticles of any composition and shape at any location in a well-defined crystalline lattice with nm-scale precision should have far-reaching implications in areas ranging from catalysis to photonics to energy.
Economic Impact
The economic impact of SNA technology is substantial and rapidly growing. Three companies have been founded that are based on SNA technology - Nanosphere, AuraSense, and AuraSense Therapeutics (founded in 2000, 2009, and 2011, respectively). These companies have employed hundreds of people and commercialized over 1800 products. Nanosphere was one of the first nanotechnology-based biotechnology firms to go public in late 2007. The FDA-cleared Verigene System has been commercialized by Nanosphere, and nine accompanying FDA-cleared panel assays (Warfarin Metabolism, F5/F2/MTHFR, Influenza A and B, Staphylococcus aureus, 2009 H1N1) are now used by hospitals all over the world. Further, hundreds of research laboratories are currently utilizing these structures in all sorts of applications spanning genomics and proteomics.
References
- 1 2 3 4 5 Cutler, J. I., et al., Spherical Nucleic Acids. J Am Chem Soc 2012, 134 (3), 1376-1391.
- ↑ Mirkin, C. A., et al., A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382 (6592), 607-609.
- ↑ Lee, J. S., et al., Silver nanoparticle-oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett 2007, 7 (7), 2112-2115.
- 1 2 Cutler, J. I., et al., Polyvalent Oligonucleotide Iron Oxide Nanoparticle "Click" Conjugates. Nano Lett 2010, 10 (4), 1477-1480.
- ↑ Xue, C., et al., Self-assembled monolayer mediated silica coating of silver triangular nanoprisms. Adv Mater 2007, 19 (22), 4071.
- 1 2 Mitchell, G. P., et al., Programmed assembly of DNA functionalized quantum dots. J Am Chem Soc 1999, 121 (35), 8122-8123.
- 1 2 Cutler, J. I., et al., Polyvalent Nucleic Acid Nanostructures. J Am Chem Soc 2011, 133 (24), 9254-9257.
- ↑ Taton, T. A., et al., Scanometric DNA array detection with nanoparticle probes. Science 2000, 289 (5485), 1757-1760.
- ↑ Prigodich, A. E., et al., Tailoring DNA Structure To Increase Target Hybridization Kinetics on Surfaces. J Am Chem Soc 2010, 132 (31), 10638-10641.
- ↑ Rosi, N. L., et al., Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science 2006, 312 (5776), 1027-1030.
- ↑ Lytton-Jean, A. K. R.; Mirkin, C. A., A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes. J Am Chem Soc 2005, 127 (37), 12754-12755.
- 1 2 3 4 Hurst, S. J., et al., "Three-Dimensional Hybridization" with polyvalent DNA-gold nanoparticle conjugates. J Am Chem Soc 2008, 130 (36), 12192-12200.
- ↑ Han, D. R., et al., DNA Origami with Complex Curvatures in Three-Dimensional Space. Science 2011, 332 (6027), 342-346.
- ↑ Seeman, N. C., DNA in a material world. Nature 2003, 421 (6921), 427-431.
- ↑ Seeman, N. C., An overview of structural DNA Nanotechnology. Mol Biotechnol 2007, 37 (3), 246-257.
- ↑ Jones, M. R., et al., DNA-nanoparticle superlattices formed from anisotropic building blocks. Nat Mater 2010, 9 (11), 913-917.
- ↑ Alivisatos, A. P., et al., Organization of 'nanocrystal molecules' using DNA. Nature 1996, 382 (6592), 609-611.
- ↑ Giljohann, D. A., et al., Gene Regulation with Polyvalent siRNA-Nanoparticle Conjugates. J Am Chem Soc 2009, 131 (6), 2072.
- ↑ Giljohann, D. A., et al., Oligonucleotide loading determines cellular uptake of DNA-modified gold nanoparticles. Nano Lett 2007, 7 (12), 3818-3821.
- ↑ Massich, M. D., et al., Regulating Immune Response Using Polyvalent Nucleic Acid-Gold Nanoparticle Conjugates. Mol Pharmaceut 2009, 6 (6), 1934-1940.
- ↑ Massich, M. D., et al., Cellular Response of Polyvalent Oligonucleotide-Gold Nanoparticle Conjugates. ACS Nano 2010, 4 (10), 5641-5646.
- ↑ Seferos, D. S., et al., Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett 2009, 9 (1), 308-311.
- ↑ Fellman, Megan (30 October 2013). "Incurable Brain Cancer Gene is Silenced". Northwestern University. Retrieved 26 November 2013.
- ↑ Jensen, Samuel; Day, Emily; Ko, Caroline; Mirkin, Chad; Stegh, Alexander; et al. (2013). "Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma". Science Translational Medicine. AAAS. 5 (209): 209ra152. doi:10.1126/scitranslmed.3006839.
- ↑ Cao, Y. W. C., et al., Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science 2002, 297 (5586), 1536-1540.
- 1 2 Georganopoulou, D. G., et al., Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer's disease. P Natl Acad Sci USA 2005, 102 (7), 2273-2276.
- ↑ Giljohann, D. A., et al., Gold Nanoparticles for Biology and Medicine. Angew Chem Int Edit 2010, 49 (19), 3280-3294.
- ↑ Nam, J. M., et al., Bio-barcodes based on oligonucleotide-modified nanoparticles. J Am Chem Soc 2002, 124 (15), 3820-3821.
- ↑ Nam, J. M., et al., Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science 2003, 301 (5641), 1884-1886.
- 1 2 Seferos, D. S., et al., Nano-flares: Probes for transfection and mRNA detection in living cells. J Am Chem Soc 2007, 129 (50), 15477.
- ↑ Thaxton, C. S., et al., Nanoparticle-based bio-barcode assay redefines "undetectable" PSA and biochemical recurrence after radical prostatectomy. P Natl Acad Sci USA 2009, 106 (44), 18437-18442.
- ↑ Kim, E. Y., et al., Detection of HIV-1 p24 Gag in plasma by a nanoparticle-based bio-barcode-amplification method. Nanomedicine-UK 2008, 3 (3), 293-303.
- ↑ Tang, S. X., et al., Nanoparticle-based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. Jaids-J Acq Imm Def 2007, 46 (2), 231-237.
- ↑ Buchan, B. W., et al., Evaluation of a Microarray-Based Genotyping Assay for the Rapid Detection of Cytochrome P450 2C19 *2 and *3 Polymorphisms From Whole Blood Using Nanoparticle Probes. Am J Clin Pathol 2011, 136 (4), 604-608.
- ↑ Lefferts, J. A., et al., Evaluation of the Nanosphere Verigene (R) System and the Verigene (R) F5/F2/MTHFR Nucleic Acid Tests. Exp Mol Pathol 2009, 87 (2), 105-108.
- ↑ Wilson, S. R., et al., Detection of myocardial injury in patients with unstable angina using a novel nanoparticle cardiac troponin I assay: Observations from the PROTECT-TIMI 30 Trial. Am Heart J 2009, 158 (3), 386-391.
- ↑ Storhoff, J., et al., Detection of prostate cancer recurrence using an ultrasensitive nanoparticle-based PSA assay. J Clin Oncol 2009, 27 (15).
- ↑ Macfarlane, R. J., et al., Nanoparticle Superlattice Engineering with DNA. Science 2011, 334 (6053), 204-208.
- ↑ Auyeung, E., et al., Synthetically programmable nanoparticle superlattices using a hollow three-dimensional spacer approach. Nat Nanotechnol 2012, 7 (1), 24-28.