Affimer
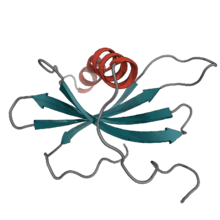
Affimer[1] molecules are small, highly stable proteins that bind their target molecules with similar specificity and affinity to that of antibodies. These engineered non-antibody binding proteins are designed to mimic the molecular recognition characteristics of monoclonal antibodies in different applications.[2] Protein engineers have attempted to improve the experimental properties of these affinity reagents, to increase their stability,[3] make them robust across a range of temperatures and pH,[4] offer small sizes of the reagent,[5] and make them easy to express at high yields in E.coli and mammalian cells.
Development
Affimer proteins were developed initially at the MRC Cancer Cell Unit in Cambridge then across two laboratories at the University of Leeds.[6][7][8][9] Derived from the cysteine protease inhibitor family of cystatins,[10] which function in nature as cysteine protease inhibitors,[11][12] these 12–14 kDa proteins share the common tertiary structure of an alpha-helix lying on top of an anti-parallel beta-sheet.[13]
Affimer proteins display two peptide loops and an N-terminal sequence that can all be randomised to bind to desired target proteins with high affinity and specificity,[3] in a similar manner to monoclonal antibodies. Stabilisation of the two peptides by the protein scaffold constrains the possible conformations that the peptides can take, increasing the binding affinity and specificity compared to libraries of free peptides.
Production
Phage display libraries of 1010 randomised potential target interaction sequences are generated and screened to identify an Affimer protein with high-specificity binding to the target protein and binding affinities in the nM range. The use of in vitro screening techniques allows affinity maturation to be performed to achieve greater binding affinities and means that the target space is not limited by an animal host’s immune system. Due to Affimer proteins being generated using recombinant systems, their generation is significantly more rapid and reproducible[14] compared to the production of traditional antibodies.
Multimeric forms of Affimer proteins have been generated and shown to yield titrimetric volumes in the range of 200–400 mg/L under small scale culture using bacterial host systems. Multimeric forms of Affimer proteins with the same target specificity provided avidity effects in target binding, while fusion of different Affimer proteins with different target specificities would enable multi-specific affinity proteins.[15]
Affimer binders have been produced to a large number of targets including ubiquitin chains,[16] immunoglobulins,[17] C-reactive protein,[18] interleukin-8,[19] complement C3[20][21] and magnetite nanoparticles[4] for use in a number of molecular recognition applications.
Many different tags and fusion proteins, such as the fluorophores, the His tag and c-Myc have been previously conjugated to Affimer proteins for use in various research applications. Specific cysteine residues were introduced to the protein to allow thiol chemistry to uniformally orient Affimer proteins on a solid support in the development of biosensors[19] and click chemistry has been used to conjugate Affimer proteins to MRI contrast reagents for the purposes of targeted imaging.[22]
Properties
Affimer binders are recombinant proteins. They display the robust characteristics of high thermostability, with melting temperatures over 80 °C,[23] resistance to extremes of pH (2–13.7),[23] freeze-thaw cycles and lyophilisation. The low molecular weight[24] of Affimer binders means that problems of steric hindrance, that are typically observed with antibodies, may be avoided.
These synthetic antibodies were engineered to be stable, non-toxic, biologically neutral and contain no post-translational modifications or disulfide bridges.[2] Two separate loop sequences, incorporating a total of 12 to 36 amino acids, form the target interaction surface so interaction surfaces can range form 650–1000 Å. The large interaction surface is purported to result in highly-specific, high affinity binding to target proteins.[6][9] As a result Affimer molecules can distinguish between proteins that differ by only a single amino acid,[25] can detect subtle changes in protein expression levels even in a multiplexed format and can distinguish between multiple closely related protein domains.[26]
Applications
Affimer binders have been used across a number of platforms, including ELISA,[27] surface plasmon resonance, affinity purification, immunohistochemistry[28] and flow cytometry. Affimer reagents that inhibit protein-protein interactions can also be produced with the potential to express these inhibitors in mammalian cells to investigate and modify signalling pathways.[29][30] They have also been co-crystallised in complex with target proteins,[31] enabling drug discovery through in silico screening and displacement assays.
Affimer technology has been commercialised and developed by Avacta Life Sciences, who are developing these affinity reagents as tools for research and diagnostics and as biotherapeutics.
References
- ↑ Proprietary name, owned by Avacta
- 1 2 The Scientist. "Antibody Alternatives".
- 1 2 Sharma R.; Deacon S.E.; Nowak D.; George S.E.; Szymonik M.P.; Tang A.A.S.; Tomlinson D.C.; Davis A.G.; McPherson M.J.; Wälti C. (2016). "Label-free electrochemical impedance biosensor to detect human interleukin-8 in serum with sub-pg/ml sensitivity.". Biosensors and Bioelectronics. 80: 607–13. doi:10.1016/j.bios.2016.02.028.
- 1 2 Rawlings A.E.; Bramble J.P.; Tang A.A.S.; Somner L.A.; Monnington A.E.; Cooke D.J.; McPherson M.J.; Tomlinson D.C.; Staniland S.S. (2015). "Phage display selected magnetite interacting adhirons for shape controlled nanoparticle synthesis". Chem. Sci. 6: 5586–94. doi:10.1039/C5SC01472G.
- ↑ Roberts, Josh P. (2013). "Biomarkers Take Center Stage". GEN. 33.
- 1 2 Woodman R.; Yeh J. T-H.; Laurenson S.; Ko Ferrigno P. (2005). "Design and validation of a neutral protein scaffold for the presentation of peptide aptamers". J. Mol. Biol. 352 (5): 1118–33. doi:10.1016/j.jmb.2005.08.001.
- ↑ Hoffmann T.; Stadler L.K.J.; Busby M.; Song Q.; Buxton A.T.; Wagner S.D.; Davis J.J.; Ko Ferrigno P. (2010). "Structure-function studies of an engineered scaffold protein derived from stefin A. I:Development of the SQT variant.". Protein Eng. Des. Sel. 23 (5): 403–13. doi:10.1093/protein/gzq012. PMC 2851446
 .
. - ↑ Stadler L.K.; Hoffmann T.; Tomlinson D.C.; Song Q.; Lee T.; Busby M.; Nyathi Y.; Gendra E.; Tiede C.; Flanagan K.; Cockell S.J.; Wipat A.; Harwood C.; Wagner S.D.; Knowles M.A.; Davis J.J.; Keegan N.; Ko Ferrigno P. (2011). "Structure-function studies of an engineered scaffold protein derived from stefin A. II: Development and applications of the SQT variant.". Protein Eng. Des. Sel. 24 (9): 751–63. doi:10.1093/protein/gzr019. PMID 21616931.
- 1 2 Tiede C.; Tang A.A.; Deacon S.E.; Mandal U.; Nettleship J.E.; Owen R.L.; George S.E.; Harrison D.J.; Owens R.J.; Tomlinson D.C.; McPherson M.J. (2014). "Adhiron: A stable and versatile peptide display scaffold for molecular recognition applications". Protein Eng. Des. Sel. 27 (5): 145–55. doi:10.1093/protein/gzu007. PMID 24668773.
- ↑ "Affimers – Next Generation Affinity Reagents". Avacta Life Sciences. Retrieved 22 May 2014.
- ↑ Turk V.; Stoka V.; Turk D. (2008). "Cystatins: Biochemical and structural properties, and medical relevance". Front Biosci. 1 (13): 5406–5420.
- ↑ Kondo H.; Abe K.; Emori Y.; Arai S. (1991). "Gene organization of oryzastatin II, a new cystatin superfamily member of plant origin, is closely related to that of oryzacystatin-I but different from those of animal cystatins". FEBS Lett. 278: 87–90. doi:10.1016/0014-5793(91)80090-p.
- ↑ Turk V. & W. Bode (1991). "The cystatins: protein inhibitors of cysteine proteinases". FEBS Lett. 285 (2): 213–219. doi:10.1016/0014-5793(91)80804-C. PMID 1855589.
- ↑ Avacta Life Sciences. "Key Benefits of Affimers - Rapid Generation".
- ↑ Avacta Life Sciences. "Multimeric Affimer binders".
- ↑ Avacta Life Sciences. "Anti-diUbiquitin K48-linkage Affimer (36-28)".
- ↑ Avacta Life Sciences. "Anti-Immunoglobulin Research Area of Affimers".
- ↑ Johnson A, Song Q, Ko Ferrigno P, Bueno PR, Davis JJ (Aug 7, 2012). "Sensitive Affimer and antibody based impedimetric label-free assays for C-reactive protein". Anal. Chem. 84 (15): 6553–60. doi:10.1021/ac300835b. PMID 22789061.
- 1 2 Sharma R, Deacon SE, Nowak D, George SE, Szymonik MP, Tang AAS, Tomlinson DC, Davies AG, McPherson MJ, Walti C (2016). "Label-free electrochemical impedance biosensor to detect human interleukin-8 in serum with sub-pg/ml sensitivity". Biosensors and Bioelectronics. 80: 607–613. doi:10.1016/j.bios.2016.02.028.
- ↑ Avacta Life Sciences. "ALS application data- Affinity purification".
- ↑ Avacta Life Sciences. "ALS application data protein-protein interactions".
- ↑ Fisher MJ, Williamson DJ, Burslem GM, Plante JP, Manfield IM, Tiede C, Ault JR, Stockley PG, Plein S, Maqbool A, Tomlinson DC, Foster R, Warriner SL, Bon RS (2015). "Trivalent Gd-DOTA reagents for modification of proteins". RSC Advan. 5 (116): 96194–96200. doi:10.1039/C5RA20359G.
- 1 2 Avacta Life Sciences. "Key benefits of Affimers - Robustness".
- ↑ Avacta Life Sciences. "Key Benefits of Affimers - Small Size".
- ↑ Avacta Life Sciences. "ALS applications data- Difficult targets".
- ↑ Avacta Life Sciences. "ALS applications data- SH2 domains".
- ↑ Avacta Life Sciences. "ALS applications data - ELISA".
- ↑ Avacta Life Sciences. "ALS applications data - IHC".
- ↑ Avacta Life Sciences. "ALS applications data - Protein-protein interactions".
- ↑ Kyle H.F.; Wickson K.F.; Stott J.; Burslem G.M.; Breeze A.L.; Tiede C.; Tomlinson D.C.; Warriner S.L.; Nelson A.; Wilson A.J.; Edwards T.A. (2015). "Exploration of the HIF-1α/p300 interface using peptide and adhiron phage display technologies". Mol. Biosyst. 11 (10): 2738–49. doi:10.1039/c5mb00284b.
- ↑ Avacta Life Sciences. "ALS application data- Allosteric inhibition of FcγRIIIa-IgG interactions".
