Digital holographic microscopy
Digital holographic microscopy (DHM) is digital holography applied to microscopy. Digital holographic microscopy distinguishes itself from other microscopy methods by not recording the projected image of the object. Instead, the light wave front information originating from the object is digitally recorded as a hologram, from which a computer calculates the object image by using a numerical reconstruction algorithm. The image forming lens in traditional microscopy is thus replaced by a computer algorithm. Other closely related microscopy methods to digital holographic microscopy are interferometric microscopy, optical coherence tomography and diffraction phase microscopy. Common to all methods is the use of a reference wave front to obtain amplitude (intensity) and phase information. The information is recorded on a digital image sensor or by a photo detector from which an image of the object is created (reconstructed) by a computer. In traditional microscopy, which do not use a reference wave front, only intensity information is recorded and essential information about the object is lost.
Digital holography has mostly been applied to light microscopy. However, digital holography has also been applied to electron microscopy.[1] Holography was invented by Dennis Gabor to improve the electron microscope. For various reasons holography never made it into the electron microscope.
Working principle
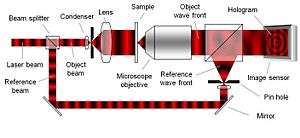
To create the necessary interference pattern, i.e., the hologram, the illumination needs to be a coherent (monochromatic) light source, a laser for example. As can be seen in Figure 2, the laser light is split into an object beam and a reference beam. The expanded object beam illuminates the sample to create the object wave front. After the object wave front is collected by a microscope objective, the object and reference wave fronts are joined by a beam splitter to interfere and create the hologram. Using the digitally recorded hologram, a computer acts as a digital lens and calculates a viewable image of the object wave front by using a numerical reconstruction algorithm.
Commonly, a microscope objective is used to collect the object wave front. However, as the microscope objective is only used to collect light and not to form an image, it may be replaced by a simple lens. If a slightly lower optical resolution is acceptable, the microscope objective may be entirely removed.
Digital holography comes in different flavors, such as off-axis Fresnel, Fourier, image plane, in-line, Gabor and phase-shifting digital holography,[2] depending on the optical setup. The basic principle, however, is the same; a hologram is recorded and an image is reconstructed by a computer.
The lateral optical resolution of digital holographic microscopy is equivalent to the resolution of traditional light microscopy. DHM is diffraction-limited by the numerical aperture, in the same way as traditional light microscopy. However, DHM offers a superb axial (depth) resolution. An axial accuracy of approximately 5 nm has been reported.[3]
Advantages
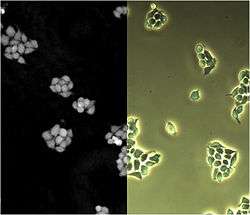
Phase shift images
Besides the ordinary bright field image, a phase shift image is created as well. The phase shift image is unique for digital holographic microscopy and gives quantifiable information about optical distance. In reflection DHM, the phase shift image forms a topography image of the object.
Transparent objects, like living biological cells, are traditionally viewed in a phase contrast microscope or in a differential interference contrast microscope. These methods visualize phase shifting transparent objects by distorting the bright field image with phase shift information. Instead of distorting the bright field image, transmission DHM creates a separate phase shift image showing the optical thickness of the object. Digital holographic microscopy thus makes it possible to visualize and quantify transparent objects and is therefore also referred to as quantitative phase contrast microscopy.
Traditional phase contrast or bright field images of living unstained biological cells, Figure 3 (right), have proved themselves to be very difficult to analyze with image analysis software. On the contrary, phase shift images, Figure 3 (left), are readily segmented and analyzed by image analysis software based on mathematical morphology, such as CellProfiler.[4]
3-Dimensional information
An object image is calculated at a given focal distance. However, as the recorded hologram contains all the necessary object wave front information, it is possible to calculate the object at any focal plane by changing the focal distance parameter in the reconstruction algorithm. In fact, the hologram contains all the information needed to calculate a complete image stack. In a DHM system, where the object wave front is recorded from multiple angles, it is possible to fully characterize the optical characteristics of the object and create tomography images of the object.[5][6]
Digital autofocus
Conventional autofocus is achieved by vertically changing the focal distance until a focused image plane is found. As the complete stack of image planes may be calculated from a single hologram, it is possible to use any passive autofocus method to digitally select the focal plane.[7] The digital auto focusing capabilities of digital holography opens up the possibility to scan and image surfaces extremely rapidly, without any vertical mechanical movement. By recording a single hologram and afterwards stitch sub-images together that are calculated at different focal planes, a complete and focused image of the object may be created.[8]
Optical aberration correction
As DHM systems do not have an image forming lens, traditional optical aberrations do not apply to DHM. Optical aberrations are "corrected" by design of the reconstruction algorithm. A reconstruction algorithm that truly models the optical setup will not suffer from optical aberrations.[9]
[10]
Low cost
In optical microscopy systems, optical aberrations are traditionally corrected by combining lenses into a complex and costly image forming microscope objective. Furthermore, the narrow focal depth at high magnifications requires precision mechanics. The needed components for a DHM system are inexpensive optics and semiconductor components, such as a laser diode and an image sensor. The low component cost in combination with the auto focusing capabilities of DHM, make it possible to manufacture DHM systems for a very low cost.[11]
[12]
Applications

Digital holographic microscopy has been successfully applied in a range of application areas.[13] However, due to DHM’s capability of non-invasively visualizing and quantifying biological tissue, bio-medical applications have received most attention.[14] Examples of bio-medical applications are:
- Label-free cell counting in adherent cell cultures. Digital holographic microscopy makes it possible to perform cell counting and to measure cell viability directly in the cell culture chamber.[15][16] Today, the most commonly used cell counting methods, hemocytometer or Coulter counter, only work with cells that are in suspension.
- Label-free viability analysis of adherent cell cultures.[17][18] Digital holography has been used to study the apoptotic process in different cell types. The refractive index changes taking place during the apoptotic process are easily measured with DHM.
- Label-free cell cycle analysis. The phase shift induced by cells has been shown to be correlated to the cell dry mass. The cell dry mass can be combined with other parameters obtainable by digital holography, such as cell volume and refractive index, to provide a better understanding of the cell cycle.[19]
- Label-free morphology analysis of cells. Digital holography has been used in different contexts to study cell morphology using neither staining nor labeling.[16] This can be used to follow processes such as the differentiation process where cell characteristics change. DHM has also been used for automated plant stem cell monitoring, and made it possible to distinguish between two types of stem cells by measuring morphological parameters.[20]
- Label free nerve cell studies. Digital holographic microscopy makes it possible to study undisturbed processes in nerve cells as no labeling is required.[21] The swelling and shape changing of nerve cells caused by cellular imbalance was easily studied.

- Label-free high content analysis. Fluorescent high content analysis/screening has several drawbacks. Label-free alternatives based on phase shift images have therefore been proposed.[4] The capability of DHM to obtain phase shift images rapidly over large areas opens up new possibilities of very rapid quantitative characterization of the cell cycle and the effects of specific pharmacological agents.
- Red blood cell analysis. Phase shift images have been used to study red blood cell dynamics.[22][23] Red blood cell volume and hemoglobin concentration has been measured by combining information from absorption and phase shift images to facilitate complete blood cell count by holographic microscopy.[24] It has furthermore been shown[25] that phase shift information discriminates immature red blood cells from mature, facilitating unstained reticulocyte count.
- Flow cytometry and particle tracking and characterization. Images created by digital holography are calculated from the recorded hologram at any time after the actual recording and at any given focal plane. By combining several images calculated from the same hologram, but at different focal planes, an increased depth of field may be obtained, which is vastly superior to what can be achieved with traditional light microscopy. The increased depth of field makes it possible to image and characterize the morphology of cells and particles while in suspension. Observations may be done directly in a microfluidic channel or statically in an observation chamber.[26][27][28]
- Time-lapse microscopy of cell division and migration.[29] The autofocus and phase shift imaging capabilities of digital holographic microscopy makes it possible to effortlessly create label-free and quantifiable time-lapse video clips of unstained cells for cell migration studies.[30] In Figure 5 a label-free time-lapse of dividing and migrating cells is shown.
- Tomography studies.[31] Digital holographic microscopy allows for label-free and quantifiable analysis of subcellular motion deep in living tissue.
History
The first reports of replacing the photographic hologram of classical holography by digitally recording the hologram and numerically reconstructing the image in a computer was published in the late 1960s[32] and in the early 1970s.[33][34] Similar ideas were proposed for the electron microscope in the early 1980s.[35] But, computers were too slow and recording capabilities were too poor for digital holography to be useful in practice. After the initial excitement, digital holography went into a similar hibernation as holography experienced about two decades earlier.
In the mid 1990s, digital image sensors and computers had become powerful enough to reconstruct images with some quality.[36] In the 1960s, digital holography could either mean to compute an image from a hologram or to compute a hologram from a 3D model. The latter developed in parallel with classical holography during the hibernation of digital holography. During that time, digital holography was synonymous with what is now known as computer generated holography.
By the mid 1990s, digital image sensors and computers had improved tremendously, but still lacked the required pixel count and density for digital holography to be anything more than a curiosity. At the time, the market driving digital image sensors was primarily low-resolution video, and so those sensors provided only PAL, NTSC, or SECAM resolution. This suddenly changed at the beginning of the 21st century with the introduction of digital still image cameras, which drove demand for inexpensive high-pixel-count sensors. As of 2010, affordable image sensors can have up to 60 megapixels. In addition, the CD and DVD-player market has driven development of affordable diode lasers and optics.
The first reports of using digital holography for light microscopy came in the mid 1990s.[37][38] However, it was not until the early 2000s that image sensor technology had progressed far enough to allow images of a reasonable quality. During this time, the first commercial digital holographic microscopy companies, 4Deep inwater imaging, Lyncée tec, and Phase Holographic Imaging, were founded. With increased computing power and use of inexpensive high-resolution sensors and lasers, digital holographic microscopy is today finding applications primarily within life science, oceanology and metrology.
See also
- digital holography
- quantitative phase contrast microscopy
- Holographic interferometry
- holography
- microscopy
References
- ↑ Martha R. McCartney; David J. Smith (2007). "Electron Holography: Phase Imaging with Nanometer Resolution". Annual Review of Materials Research. 37: 729–767. Bibcode:2007AnRMS..37..729M. doi:10.1146/annurev.matsci.37.052506.084219.
- ↑ Myung K. Kim (2010). "Principles and techniques of digital holographic microscopy". SPIE Reviews. 1: 018005. Bibcode:2010SPIER...1a8005K. doi:10.1117/6.0000006.
- ↑ Bjorn Kemper; Patrik Langehanenberg; Gert von Bally (2007). "Digital Holographic Microscopy: A New Method for Surface Analysis and Marker?Free Dynamic Life Cell Imaging" (PDF). Optik & Photonik (2): 41–44.
- 1 2 Jyrki Selinummi; Pekka Ruusuvuori; Irina Podolsky; Adrian Ozinsky; Elizabeth Gold; Olli Yli-Harja; Alan Aderem; Ilya Shmulevich (2009). Serrano-Gotarredona, Teresa, ed. "Bright Field Microscopy as an Alternative to Whole Cell Fluorescence in Automated Analysis of Macrophage Images". PLoS ONE. 4 (10): e7497. Bibcode:2009PLoSO...4.7497S. doi:10.1371/journal.pone.0007497. PMC 2760782
 . PMID 19847301.
. PMID 19847301. - ↑ Florian Charrière; Nicolas Pavillon; Tristan Colomb; Christian Depeursinge; Thierry J. Heger; Edward A. D. Mitchell; Pierre Marquet; Benjamin Rappaz (2006). "Living specimen tomography by digital holographic microscopy: morphometry of testate amoeba". Opt. Express. 14 (16): 7005–7013. Bibcode:2006OExpr..14.7005C. doi:10.1364/OE.14.007005. PMID 19529071.
- ↑ Yongjin Sung; Wonshik Choi; Christopher Fang-Yen; Kamran Badizadegan; Ramachandra R. Dasari; Michael S. Feld (2009). "Optical diffraction tomography for high resolution live cell imaging". Opt. Express. 17 (1): 266–277. Bibcode:2009OExpr..17..266S. doi:10.1364/OE.17.000266. PMC 2832333
 . PMID 19129896.
. PMID 19129896. - ↑ Frank Dubois; Cédric Schockaert; Natcaha Callens; Catherine Yourassowsky (2006). "Focus plane detection criteria in digital holography microscopy by amplitude analysis". Opt. Express. 14 (13): 5895–5908. Bibcode:2006OExpr..14.5895D. doi:10.1364/OE.14.005895. PMID 19516759.
- ↑ P. Ferraro; S. Grilli; D. Alfieri; S. De Nicola; A. Finizio; G. Pierattini; B. Javidi; G. Coppola; V. Striano (2005). "Extended focused image in microscopy by digital holography". Opt. Express. 13 (18): 6738–6749. Bibcode:2005OExpr..13.6738F. doi:10.1364/OPEX.13.006738. PMID 19498690.
- ↑ Alexander Stadelmaier; Jürgen H. Massig (2000). "Compensation of lens aberrations in digital holography". Opt. Lett. 25 (22): 1630–1632. Bibcode:2000OptL...25.1630S. doi:10.1364/OL.25.001630. PMID 18066297.
- ↑ T. Colomb; F. Montfort; J. Kühn; N. Aspert; E. Cuche; A. Marian; F. Charrière; S. Bourquin; P. Marquet; C. Depeursinge (2006). "Numerical parametric lens for shifting, magnification and complete aberration compensation in digital holographic microscopy". Journal of the Optical Society of America A. 23 (12): 3177–3190. Bibcode:2006JOSAA..23.3177C. doi:10.1364/JOSAA.23.003177.
- ↑ Aydogan Ozcan; Serhan Isikman; Onur Mudanyali; Derek Tseng; Ikbal Sencan (2010). "Lensfree on-chip holography facilitates novel microscopy applications". SPIE Newsroom. doi:10.1117/2.1201005.002947.
- ↑ Myungjun Lee; Oguzhan Yaglidere; Aydogan Ozcan (2011). "Field-portable reflection and transmission microscopy based on lensless holography". Biomedical Optics Express. 2: 2721–2730. doi:10.1364/BOE.2.002721.
- ↑ Tristan Colomb; Pierre Marquet; Florian Charrière; Jonas Kühn; Pascal Jourdain; Christian Depeursinge; Benjamin Rappaz; Pierre Magistretti (2007). "Enhancing the performance of digital holographic microscopy". SPIE Newsroom. doi:10.1117/2.1200709.0872.
- ↑ Myung-K. Kim (2010). "Applications of Digital Holography in Biomedical Microscopy". J. Opt. Soc. Korea. 14 (2): 77–89. doi:10.3807/JOSK.2010.14.2.077.
- ↑ Daniel Carl; Björn Kemper; Günther Wernicke; Gert von Bally (2004). "Parameter-Optimized Digital Holographic Microscope for High-Resolution Living-Cell Analysis". Applied Optics. 43 (33): 6536–6544. Bibcode:2004ApOpt..43.6536C. doi:10.1364/AO.43.006536.
- 1 2 Mölder A; Sebesta M; Gustafsson M; Gisselson L; Wingren AG; Alm K. (2008). "Non-invasive, label-free cell counting and quantitative analysis of adherent cells using digital holography". J Microsc. 232 (2): 240–247. doi:10.1111/j.1365-2818.2008.02095.x. PMID 19017223.
- ↑ Kemper B; Carl D; Schnekenburger J; Bredebusch I; Schäfer M; Domschke W; von Bally G (2006). "Investigations on living pancreas tumor cells by digital holographic microscopy". J. Biomed. Opt. 11 (3): 034005. Bibcode:2006JBO....11c4005K. doi:10.1117/1.2204609.
- ↑ Kemmler M; Fratz M; Giel D; Saum N; Brandenburg A; Hoffman C (2007). "Noninvasive time-dependent cytometry monitoring by digital holography". J. Biomed. Opt. 12 (6): 064002. Bibcode:2007JBO....12f4002K. doi:10.1117/1.2804926. PMID 18163818.
- ↑ Benjamin Rappaz; Elena Cano; Tristan Colomb; Jonas Kühn; Christian Depeursinge; Viesturs Simanis; Pierre J. Magistretti; Pierre Marquet (2009). "Noninvasive characterization of the fission yeast cell cycle by monitoring dry mass with digital holographic microscopy" (PDF). J. Biomed. Opt. 14 (3): 034049. Bibcode:2009JBO....14c4049R. doi:10.1117/1.3147385. PMID 19566341.
- ↑ Inkyu Moon; Bahram Javidi (2007). "Three-dimensional identification of stem cells by computational holographic imaging". J. R. Soc. Interface. 4 (13): 305–313. doi:10.1098/rsif.2006.0175. PMC 2359842
 . PMID 17251147.
. PMID 17251147. - ↑ Nicolas Pavillon; Alexander Benke; Daniel Boss; Corinne Moratal; Jonas Kühn; Pascal Jourdain; Christian Depeursinge; Pierre J. Magistretti; Pierre Marquet (2010). "Cell morphology and intracellular ionic homeostasis explored with a multimodal approach combining epifluorescence and digital holographic microscopy". J. of Biophotonics. 3 (7): 432–436. doi:10.1002/jbio.201000018.
- ↑ Gabriel Popescu; YoungKeun Park; Wonshik Choi; Ramachandra R. Dasari; Michael S. Feld; Kamran Badizadegan (2008). "Imaging red blood cell dynamics by quantitative phase microscopy" (PDF). Blood Cells, Molecules, and Diseases. 41 (1): 10–16. doi:10.1016/j.bcmd.2008.01.010.
- ↑ Marquet P.; Rappaz B.; Barbul A.; Korenstein R.; Depeursinge C.; Magistretti P. (2009). "Red blood cell structure and dynamics explored with digital holographic microscopy". Proc. Of SPIE. 7182: 71821A. doi:10.1117/12.809224.
- ↑ Mustafa Mir; et al. (2011). "Blood testing at the single cell level using quantitative phase and amplitude microscopy". Biomed. Opt. Express. 2 (12): 3259–3266. doi:10.1364/BOE.2.003259.
- ↑ Mona Mihailescu; et al. (2011). "Automated imaging, identification, and counting of similar cells from digital hologram reconstructions". Appl. Opt. 50 (20): 3589–3597. Bibcode:2011ApOpt..50.3589M. doi:10.1364/AO.50.003589.
- ↑ Fook Chiong Cheong; Bo Sun; Rémi Dreyfus; Jesse Amato-Grill; Ke Xiao; Lisa Dixon; David G. Grier (2009). "Flow visualization and flow cytometry with holographic video microscopy". Optics Express. 17 (15): 13071–13079. Bibcode:2009OExpr..1713071C. doi:10.1364/OE.17.013071.
- ↑ Shigeru Murata; Norifumi Yasuda (2000). "Potential of digital holography in particle measurement". Opt. Laser Eng. 32 (7-8): 567–574. Bibcode:2000OptLT..32..567M. doi:10.1016/S0030-3992(00)00088-8.
- ↑ Emmanouil Darakis; Taslima Khanam; Arvind Rajendran; Vinay Kariwala; Thomas J. Naughton; Anand K. Asundi (2010). "Microparticle characterization using digital holography". Chem. Eng. Sci. 65 (2): 1037–1044. doi:10.1016/j.ces.2009.09.057.
- ↑ Björn Kemper; Andreas Bauwens; Angelika Vollmer; Steffi Ketelhut; Patrik Langehanenberg (2010). "Label-free quantitative cell division monitoring of endothelial cells by digital holographic microscopy". J. Biomed. Opt. 15 (3): 036009. Bibcode:2010JBO....15c6009K. doi:10.1117/1.3431712. PMID 20615011.
- ↑ Johan Persson; Anna Mölder; Sven-Göran Pettersson; Kersti Alm (2010). "Cell motility studies using digital holographic microscopy". In A. Méndez-Vilas and J. Díaz. Microscopy: Science, Technology, Applications and Education (PDF). Microscopy Series Nº 4. 2. FORMATEX. pp. 1063–1072.
- ↑ Kwan Jeong; John J. Turek; David D. Nolte (2007). "Fourier-domain digital holographic optical coherence imaging of living tissue". Appl. Opt. 46 (22): 4999–5008. Bibcode:2007ApOpt..46.4999J. doi:10.1364/AO.46.004999. PMID 17676107.
- ↑ Goodman J. W.; Lawrence R. W. (1967). "Digital image formation from electronically detected holograms". Appl. Phys. Lett. 11 (3): 77–79. Bibcode:1967ApPhL..11...77G. doi:10.1063/1.1755043.
- ↑ Huang T. (1971). "Digital Holography". Proc. Of IEEE. 59 (9): 1335–1346. doi:10.1109/PROC.1971.8408.
- ↑ Kronrod M. A.; Merzlyakov N. S.; Yaroslavskii L. P. (1972). "Reconstruction of holograms with a computer". Sov. Phys. Tech. Phys. 17: 333–334. Bibcode:1972SPTP...17..333K.
- ↑ Cowley J. M; Walker D. J. (1981). "Reconstruction from in-line holograms by digital processing". Ultramicroscopy. 6: 71–76. doi:10.1016/S0304-3991(81)80179-9.
- ↑ Schnars U.; Jüptner W. (1994). "Direct recording of holograms by a CCD target and numerical reconstruction". Applied Optics. 33 (2): 179–181. Bibcode:1994ApOpt..33..179S. doi:10.1364/AO.33.000179. PMID 20862006.
- ↑ Cuche E.; Poscio P.; Depeursinge C. (1996). "Optical tomography at the microscopic scale by means of a numerical". Proc. SPIE. 2927: 61. doi:10.1117/12.260653.
- ↑ Tong Zhang; Ichirou Yamaguchi (1998). "Three-dimensional microscopy with phase-shifting digital holography". Optics Letters. 23 (15): 1221–1223. Bibcode:1998OptL...23.1221Z. doi:10.1364/OL.23.001221. PMID 18087480.
External links
- Digital Holography and Three Dimensional Imaging Meeting by The Optical Society
- Submersible Inline Digital Holographic Microscope by 4Deep inwater imaging
- Digital Holographic Microscopy by LyncéeTec, pioneer in digital holography
- Digital Holographic Microscopy by Phase Holographic Imaging
- Holovibes Real-time hologram rendering software
Further reading
Books
- Methods of digital holography by L. P. Yaroslavskii and N. S. Merzlyakov, Springer (1980)
- Digital Holography and Digital Image Processing: Principles, Methods, Algorithms by Leonid Yaroslavsky, Kluwer (2004)
- Handbook of Holographic Interferometry: Optical and Digital Methods by Thomas Kreis, Wiley (2004)
- Digital Holography by U. Schnars and W. Jueptner, Springer (2005)
- Digital Holography and Three-Dimensional Display: Principles and Applications by Ting-Chung Poon (Editor), Springer (2006)
- Digital Holography Microscopy applications: Three Dimensional Object Analysis and Tracking by Cedric Schockaert, VDM Verlag (2009)
- Holographic Microscopy of Phase Microscopic Objects: Theory and Practice by Tatyana Tishko, Tishko Dmitry, Titar Vladimir, World Scientific (2010)
- Quantitative Phase Imaging of Cells and Tissues by Gabriel Popescu, McGraw-Hill (2011)
- Digital Holographic Microscopy: Principles, Techniques and Applications by Myung K. Kim, Springer (2011)
- Coherent Light Microscopy: Imaging and Quantitative Phase Analysis edited by Pietro Ferraro, Springer (2011)
- Holography, Research and Technologies edited by Joseph Rosen, InTech (2011)
- Digital Holography for MEMS and Microsystem Metrology edited by Erdal Cayirci, Wiley (2011)
- Image Processing For Digital Holography by Karen Molony, VDM Verlag (2011)
- Digital Holography by Pascal Picart and Jun-chang Li, Wiley (2012)
Reviews
- General: Principles and techniques of digital holographic microscopy by Myung K. Kim, SPIE Reviews, Vol. 1, 018005 (2010)
- Cell biology: Digital Holography and Cell Studies by Kersti Alm et al., InTech (2011)
Feature issues
- DIGITAL HOLOGRAPHY AND 3-D IMAGING edited by Ting-Chung Poon, Applied Optics Vol. 50, Iss. 34 (2011)