SENP1
| View/Edit Human | View/Edit Mouse |
Sentrin-specific protease 1 is an enzyme that in humans is encoded by the SENP1 gene.[3][4][5]
General
So far there are seven SUMO proteases in humans that have been designated SENP1-7 (sentrin/SUMO-specific protease).1 The seven proteases possess a conserved C-terminal domain which are variable in size, and with a distinct N-terminal domain between them. The C-terminal domain shows catalytic activity and N-terminal domain regulates cell localization and substrate specificity.[6]
Features
SENP1 (Sentrin-specific protease 1) is a human protease of 643 amino acids with a weight of 73 kDa, EC number in humans 3.4.22.B70, which adopts a conformation that identifies it as a member of the superfamily of cysteine proteases contain a catalytic triad with characterized three amino acids: a cysteine at position 602, a histidine at position 533 and aspartic acid at position 550. The important nucleophile is cysteine located at the N-terminal alpha helix of the protein core, the other two amino acids, aspartate and histidine, are located in a beta sheet end. [7]
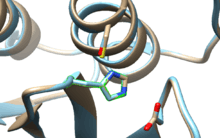
Location
Both SENP1 are located in the nucleus and cytosol depending on the cell type, although it has been seen that is exported out from the nucleus to the cytosol through a sequence of nuclear export (NES) that is located at the C-terminus. The mammalian SENP1 is localized mainly in the nucleus.[8]
Function
SENP1 catalyzes maturation SUMO protein (small ubiquitin-related modifier), which causes hydrolysis peptide bond of SUMO is in a conserved sequence Gly-Gly-|-Ala-Thr-Tyr at the C-terminal [9] to be added to the conjugation of other proteins (sumoylation).[10] In vertebrates there are three members of the family of SUMO: SUMO-1, -2 and -3. SENP1 can catalyze any of these three. This conjugation of SUMO toward other proteins is a lot like ubiquitination, however these modifications leads to different results depending on the type of protein been modified.[11]
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Gong L, Millas S, Maul GG, Yeh ET (Feb 2000). "Differential regulation of sentrinized proteins by a novel sentrin-specific protease". The Journal of Biological Chemistry. 275 (5): 3355–9. doi:10.1074/jbc.275.5.3355. PMID 10652325.
- ↑ Bailey D, O'Hare P (Jan 2004). "Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1". The Journal of Biological Chemistry. 279 (1): 692–703. doi:10.1074/jbc.M306195200. PMID 14563852.
- ↑ "Entrez Gene: SENP1 SUMO1/sentrin specific peptidase 1".
- ↑ Xu Z, Chau SF, Lam KH, Chan HY, Ng TB, Au SW (Sep 2006). "Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease". The Biochemical Journal. 398 (3): 345–52. doi:10.1042/BJ20060526. PMC 1559472
 . PMID 16712526.
. PMID 16712526. - ↑ Shen LN, Dong C, Liu H, Naismith JH, Hay RT (2006). "The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing". The Biochemical Journal. 397 (2): 279–288. doi:10.1042/BJ20052030. PMC 1513277
 . PMID 16553580.
. PMID 16553580. - ↑ Kim YH, Sung KS, Lee SJ, Kim YO, Choi CY, Kim Y (2005). "Desumoylation of homeodomain-interacting protein kinase 2 (HIPK2) through the cytoplasmic-nuclear shuttling of the SUMO-specific protease SENP1". FEBS Letters. 579 (27): 6272–6278. doi:10.1016/j.febslet.2005.10.010. PMID 16253240.
- ↑ http://www.uniprot.org/uniprot/Q9P0U3#ptm_processing
- ↑ Xu Z, Au SW (2005). "Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1". The Biochemical Journal. 386 (Pt 2): 325–330. doi:10.1042/BJ20041210. PMC 1134797
 . PMID 15487983.
. PMID 15487983. - ↑ Xu Z, Chau SF, Lam KH, Chan HY, Ng TB, Au SW (2006). "Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease". The Biochemical Journal. 398 (3): 345–52. doi:10.1042/BJ20060526. PMC 1559472
 . PMID 16712526.
. PMID 16712526.
Further reading
- Mikolajczyk J, Drag M, Békés M, Cao JT, Ronai Z, Salvesen GS (Sep 2007). "Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs". The Journal of Biological Chemistry. 282 (36): 26217–24. doi:10.1074/jbc.M702444200. PMID 17591783.
- Drag M, Mikolajczyk J, Krishnakumar IM, Huang Z, Salvesen GS (Jan 2008). "Activity profiling of human deSUMOylating enzymes (SENPs) with synthetic substrates suggests an unexpected specificity of two newly characterized members of the family". The Biochemical Journal. 409 (2): 461–9. doi:10.1042/BJ20070940. PMID 17916063.
- Cheng J, Bawa T, Lee P, Gong L, Yeh ET (Aug 2006). "Role of desumoylation in the development of prostate cancer". Neoplasia. 8 (8): 667–76. doi:10.1593/neo.06445. PMC 1601940
 . PMID 16925949.
. PMID 16925949. - Bailey D, O'Hare P (Dec 2002). "Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease". The Journal of General Virology. 83 (Pt 12): 2951–64. doi:10.1099/0022-1317-83-12-2951. PMID 12466471.
- Cheng J, Wang D, Wang Z, Yeh ET (Jul 2004). "SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1". Molecular and Cellular Biology. 24 (13): 6021–8. doi:10.1128/MCB.24.13.6021-6028.2004. PMC 480885
 . PMID 15199155.
. PMID 15199155. - Xu Z, Au SW (Mar 2005). "Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1". The Biochemical Journal. 386 (Pt 2): 325–30. doi:10.1042/BJ20041210. PMC 1134797
 . PMID 15487983.
. PMID 15487983. - Boggio R, Colombo R, Hay RT, Draetta GF, Chiocca S (Nov 2004). "A mechanism for inhibiting the SUMO pathway". Molecular Cell. 16 (4): 549–61. doi:10.1016/j.molcel.2004.11.007. PMID 15546615.
- Cheng J, Perkins ND, Yeh ET (Apr 2005). "Differential regulation of c-Jun-dependent transcription by SUMO-specific proteases". The Journal of Biological Chemistry. 280 (15): 14492–8. doi:10.1074/jbc.M412185200. PMID 15701643.
- Rajan S, Plant LD, Rabin ML, Butler MH, Goldstein SA (Apr 2005). "Sumoylation silences the plasma membrane leak K+ channel K2P1". Cell. 121 (1): 37–47. doi:10.1016/j.cell.2005.01.019. PMID 15820677.
- Veltman IM, Vreede LA, Cheng J, Looijenga LH, Janssen B, Schoenmakers EF, Yeh ET, van Kessel AG (Jul 2005). "Fusion of the SUMO/Sentrin-specific protease 1 gene SENP1 and the embryonic polarity-related mesoderm development gene MESDC2 in a patient with an infantile teratoma and a constitutional t(12;15)(q13;q25)". Human Molecular Genetics. 14 (14): 1955–63. doi:10.1093/hmg/ddi200. PMID 15917269.
- Kim YH, Sung KS, Lee SJ, Kim YO, Choi CY, Kim Y (Nov 2005). "Desumoylation of homeodomain-interacting protein kinase 2 (HIPK2) through the cytoplasmic-nuclear shuttling of the SUMO-specific protease SENP1". FEBS Letters. 579 (27): 6272–8. doi:10.1016/j.febslet.2005.10.010. PMID 16253240.
- Shen LN, Dong C, Liu H, Naismith JH, Hay RT (Jul 2006). "The structure of SENP1-SUMO-2 complex suggests a structural basis for discrimination between SUMO paralogues during processing". The Biochemical Journal. 397 (2): 279–88. doi:10.1042/BJ20052030. PMC 1513277
 . PMID 16553580.
. PMID 16553580. - Xu Z, Chau SF, Lam KH, Chan HY, Ng TB, Au SW (Sep 2006). "Crystal structure of the SENP1 mutant C603S-SUMO complex reveals the hydrolytic mechanism of SUMO-specific protease". The Biochemical Journal. 398 (3): 345–52. doi:10.1042/BJ20060526. PMC 1559472
 . PMID 16712526.
. PMID 16712526. - Shen L, Tatham MH, Dong C, Zagórska A, Naismith JH, Hay RT (Dec 2006). "SUMO protease SENP1 induces isomerization of the scissile peptide bond". Nature Structural & Molecular Biology. 13 (12): 1069–77. doi:10.1038/nsmb1172. PMC 3326531
 . PMID 17099698.
. PMID 17099698.